| Line 17: | Line 17: | ||
<h1 class="heavy">EXPERIMENTS</h1> | <h1 class="heavy">EXPERIMENTS</h1> | ||
<hr/> | <hr/> | ||
| − | <p> Here, you find all the necessary information about the wetlab materials and methods we used in the course of our project.</p> | + | <p> Here, you will find all the necessary information about the wetlab materials and methods we used in the course of our project.</p> |
<!-- Menu Generalissime --> | <!-- Menu Generalissime --> | ||
<div class="accordion" id="accordionExperiment"> | <div class="accordion" id="accordionExperiment"> | ||
| Line 48: | Line 48: | ||
<h5 class="mb-0"> | <h5 class="mb-0"> | ||
| − | <h2> | + | <h2>Genetic</h2> |
</h5> | </h5> | ||
| Line 59: | Line 59: | ||
<h3><em>Escherichia coli</em> vectors</h3> | <h3><em>Escherichia coli</em> vectors</h3> | ||
<h4>pET28</h4> | <h4>pET28</h4> | ||
| − | <p>The pET28 vector contains two His Tags in | + | <p>The pET28 vector contains two His Tags in its MCS and is specified for protein expression at high yield. It gives the strain a resistance to Kanamycin and its replication origin is the pBR322 which is a mid - low copy number plasmid (~10). The vector used was a kind gift from IBCG library. </p> |
<div class="center"> | <div class="center"> | ||
<figure class="figure"> | <figure class="figure"> | ||
| Line 66: | Line 66: | ||
</figure></div>& | </figure></div>& | ||
<h4>pETDuet-1</h4> | <h4>pETDuet-1</h4> | ||
| − | <p>The pETDuet-1 is, | + | <p>The pETDuet-1 is, like the pET28, specialized in high yield protein production. Its main difference is that it contains two MCS and can therefore express two proteins at the same time under the same expression conditions. It bears ampicilin resistance and is a mid - low copy number plasmid. The plasmid used was a gift from IBCG library.</p> |
<div class="center"> | <div class="center"> | ||
<figure class="figure"> | <figure class="figure"> | ||
| Line 74: | Line 74: | ||
<h3><em>Pichia pastoris</em> vectors</h3> | <h3><em>Pichia pastoris</em> vectors</h3> | ||
<h4>pPICZ alpha</h4> | <h4>pPICZ alpha</h4> | ||
| − | <p>The pPICZalpha | + | <p>The pPICZalpha is an integrative plasmid for <i>Pichia pastoris</i> protein expression and suitable for <i>E. coli</i> replication. Its MCS is suitable for protein expession using the methanol inductible AOX1 Pomoter (PAOX1) and offering the alpha factor secretion signal for protein purification from culture supernatant. It contains a His tag and a Myc Tag for affinity purification stategies.</p> |
<div class="center"> | <div class="center"> | ||
<figure class="figure"> | <figure class="figure"> | ||
| Line 81: | Line 81: | ||
</figure></div> | </figure></div> | ||
<h4>pGAPZalpha </h4> | <h4>pGAPZalpha </h4> | ||
| − | <p>The pGAPZalpha is roughly the same as the pPICZalpha with | + | <p>The pGAPZalpha is roughly the same as the pPICZalpha with a different promoter that is the GAP Promoter, a glucose inductible promoter.</p> |
<div class="center"> | <div class="center"> | ||
<figure class="figure"> | <figure class="figure"> | ||
| Line 258: | Line 258: | ||
</table><p>** Only for solid medium.</p> | </table><p>** Only for solid medium.</p> | ||
| − | <p><em>Nb</em>: For culture with zeocin as an antibiotic, the NaCl concentration | + | <p><em>Nb</em>: For culture with zeocin as an antibiotic, the NaCl concentration has to be 4g/L.</p> |
<h3 id="hs-medium" class="heavy">HS Medium</h3><hr/> | <h3 id="hs-medium" class="heavy">HS Medium</h3><hr/> | ||
<hr> | <hr> | ||
| Line 327: | Line 327: | ||
</tbody> | </tbody> | ||
</table><p>** Only for solid medium</p> | </table><p>** Only for solid medium</p> | ||
| − | <p><em>NB</em>: Autoclave | + | <p><em>NB</em>: Autoclave the glucose solution separately from the rest.<br> |
Autoclaving glucose separately from amino acids avoids Maillard reaction, which can result in the formation of toxic byproducts in the media.</p> | Autoclaving glucose separately from amino acids avoids Maillard reaction, which can result in the formation of toxic byproducts in the media.</p> | ||
<h3 id="tb-medium">TB medium</h3><hr/> | <h3 id="tb-medium">TB medium</h3><hr/> | ||
| Line 354: | Line 354: | ||
</tbody> | </tbody> | ||
| − | </table><p>For solid media, add 15g of agarose | + | </table><p>For solid media, add 15g of agarose per liter.<br> |
Autoclave : 20min at 121°C</p> | Autoclave : 20min at 121°C</p> | ||
| Line 371: | Line 371: | ||
<li>Dissolve 69 g YNB (without aminoacids; with ammonium sulfat) in 500 mL bidest water and filter sterilize.</li> | <li>Dissolve 69 g YNB (without aminoacids; with ammonium sulfat) in 500 mL bidest water and filter sterilize.</li> | ||
<li>Store at 4 °C.</li> | <li>Store at 4 °C.</li> | ||
| − | <li> | + | <li>Will last for one year.</li> |
</ul> | </ul> | ||
<p><strong>Biotin 500X stock solution</strong>:</p> | <p><strong>Biotin 500X stock solution</strong>:</p> | ||
| Line 446: | Line 446: | ||
<tr> | <tr> | ||
<td align="center">Glycerol</td> | <td align="center">Glycerol</td> | ||
| − | <td>1 | + | <td>1%</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 672: | Line 672: | ||
- High-copy plasmid : 1-3 ml<br> | - High-copy plasmid : 1-3 ml<br> | ||
- Low-copy plasmid : 1-5 ml<br> | - Low-copy plasmid : 1-5 ml<br> | ||
| − | Pellet 1-5ml of an overnight recombinant <i>E. coli</i> culture by | + | Pellet 1-5ml of an overnight recombinant <i>E. coli</i> culture by centrifuging at <strong>4,500rpm</strong> for <strong>4min</strong>. Discard the supernatant.</li> |
<li><em>Resuspend cells</em><br> | <li><em>Resuspend cells</em><br> | ||
| − | Completely resuspend the bacterial pellet with <strong>200 µl</strong> of the Resuspension Solution. Vortex or pipette up and down to thoroughly the cells until homogeneous.</li> | + | Completely resuspend the bacterial pellet with <strong>200 µl</strong> of the Resuspension Solution. Vortex or pipette up and down to thoroughly mix the cells until homogeneous.</li> |
<li><em>Lyse cells</em><br> | <li><em>Lyse cells</em><br> | ||
| − | Lyse the resuspended cells by adding <strong>200 µl</strong> of the lysis Solution. Immediately mix the contents by | + | Lyse the resuspended cells by adding <strong>200 µl</strong> of the lysis Solution. Immediately mix the contents by gentle inversion (6-8 times) until the mixture becomes clear and viscous.<br> |
<em>NB</em> : Do not allow the lysis reaction to exceed 5 minutes.</li> | <em>NB</em> : Do not allow the lysis reaction to exceed 5 minutes.</li> | ||
<li><em>Neutralize</em><br> | <li><em>Neutralize</em><br> | ||
| Line 859: | Line 859: | ||
- 1.0M Sorbitol<br> | - 1.0M Sorbitol<br> | ||
- 1.5ml tubes<br> | - 1.5ml tubes<br> | ||
| − | - | + | - Sterile water (1.250l)<br> |
- Centrifuge (with temperature control)</p> | - Centrifuge (with temperature control)</p> | ||
<p><strong>Protocol</strong></p> | <p><strong>Protocol</strong></p> | ||
<ol> | <ol> | ||
| − | <li>Grow 500ml cells to DO 1.3 to 1.5 at 600 nm and <strong>keep in | + | <li>Grow 500ml cells to DO 1.3 to 1.5 at 600 nm and <strong>keep in ice for the duration of the procedure)</strong></li> |
<li>Centrifuge cells for 5 min (all centrifugation steps were at 4000g at 8°C)</li> | <li>Centrifuge cells for 5 min (all centrifugation steps were at 4000g at 8°C)</li> | ||
<li>Add 100ml YPD/0.02M HEPES</li> | <li>Add 100ml YPD/0.02M HEPES</li> | ||
Revision as of 00:31, 17 October 2018
EXPERIMENTS
Here, you will find all the necessary information about the wetlab materials and methods we used in the course of our project.
Escherichia coli vectors
pET28
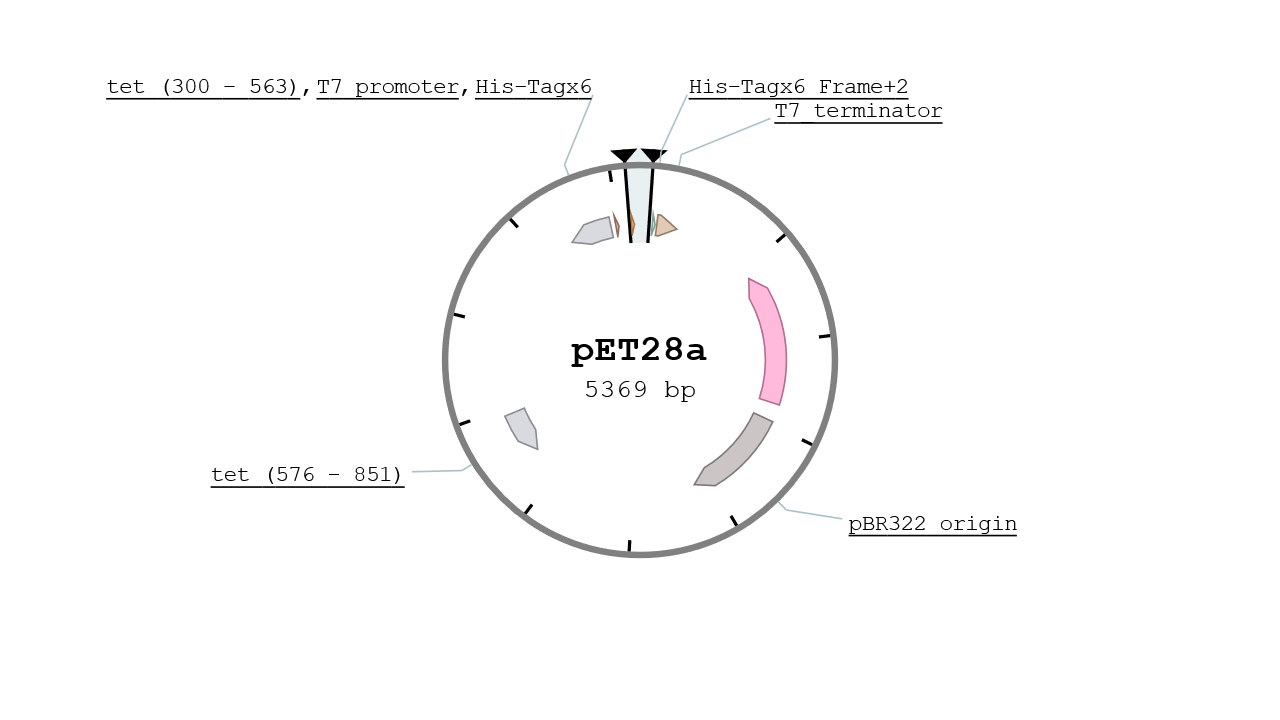
The pET28 vector contains two His Tags in its MCS and is specified for protein expression at high yield. It gives the strain a resistance to Kanamycin and its replication origin is the pBR322 which is a mid - low copy number plasmid (~10). The vector used was a kind gift from IBCG library.
pETDuet-1
The pETDuet-1 is, like the pET28, specialized in high yield protein production. Its main difference is that it contains two MCS and can therefore express two proteins at the same time under the same expression conditions. It bears ampicilin resistance and is a mid - low copy number plasmid. The plasmid used was a gift from IBCG library.

Pichia pastoris vectors
pPICZ alpha
The pPICZalpha is an integrative plasmid for Pichia pastoris protein expression and suitable for E. coli replication. Its MCS is suitable for protein expession using the methanol inductible AOX1 Pomoter (PAOX1) and offering the alpha factor secretion signal for protein purification from culture supernatant. It contains a His tag and a Myc Tag for affinity purification stategies.

pGAPZalpha
The pGAPZalpha is roughly the same as the pPICZalpha with a different promoter that is the GAP Promoter, a glucose inductible promoter.

Primer used
- Cerberus Forward : TAAGAAGGAGATATACCATGGCGGAAGCGGGTATCACC
- Cerberus Reverse : CTCGAGTGCGGCCGCAAGCTTCGGATCGTCCTATGATGGAGG
- Sirius Forward: TAAGAAGGAGATATACCATGAATGCTACGCCAACTAAGGGTGC
- Sirius Reverse: CTCGAGTGCGGCCGCAAGCTTAGCACCGGTGGAGTGACG
- BirA Forward: AAGGAGATATACATATGAAGGATAACACCGTGCCACTGA
- BirA Reverse: CTTTACCAGACTCGATTATTTTTCTGCACTACGCAGGGA
- BFP Forward: ACCACAGCCAGGATCCTATGAGCGAACTGATCAAAGAGAACA
- BFP Reverse: ATGCGGCCGCAAGCTTCTCATGCCATTCAATTTTCTGTGCT
- RFP Forward: AGGAGATATACCATGGCTTCCTCCGAAGACGTTATCAAAG
- RFP Reverse: GTGCGGCCGCAAGCTTAGCACCGGTGGAGTGACG
- Scygonadin Forward: GGCTGAAGCTGAATTCGGCCAGGCACTCAACAAAC
- Scygonadin Reverse: TGGGCCACGTGAATTCTCACTCATGCCATTCAATTTTCTG
Escherichia coli strains
Plasmid amplification strains- Stellar F-, endA1, supE44, thi-1, recA1, relA1, gyrA96, phoA, Φ80d lacZΔ M15, Δ(lacZYA-argF) U169, Δ(mrr-hsdRMS-mcrBC), ΔmcrA, λ-
- Top10 F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ-
- BL21(DE3) B F-ompT gal dcm lon hsdSB(rB-mB-) λ(DE3[lacI lacUV5-T7p07 ind1 sam7 nin5])[malB+]K-12(λS)
- Tuner F- ompT hsdSB (rB- mB-) gal dcm lacY1(DE3)
Pichia pastoris strains
For protein production we used GS200(ΔARG4, ΔHIS4) Strain for Cerberus production and X33(wt) for Scygonadin
Gluconacetobacter hansenii strain ATCC 53582
Click reagants
- DBCO-Biotin, Sigma CAS : 1255942-07-4
- DBCO-Fluorescein, Jena Bioscience Cat. No. : CLK-051-1
- DBCO-Magnetic beads, Jena Bioscience Cat. No. : CLK-1037-1
- Fluorescein-Azide, Jena Bioscience Cat. No. : CLK-80101-5
- 4-L-azidophenylanaline, Sigma Ref. : 06162
Graphene Functionalization
- Graphene, Sigma Ref. : 900561
- 4-Ethynylaniline, Sigma Ref. : 481122
- Isopentyl Nitrite, Sigma Ref. : 150495
- N,N-Dimathylformamide, Sigma Ref. : 227056
Protein purification
- Cobalt Affinity Gel, Sigma Ref. : H8162
- Protease inhibitor cocktail tablet, Sigma Ref. : S8830
Cellulose Binding assay
- Avicel, Sigma Ref. : 11365
No dogs were harmed over the course of this iGEM project.
The whole Toulouse INSA-UPS team wants to thank our sponsors, especially:









And many more. For futher information about our sponsors, please consult our Sponsors page.
The content provided on this website is the fruit of the work of the Toulouse INSA-UPS iGEM Team. As a deliverable for the iGEM Competition, it falls under the Creative Commons Attribution 4.0. Thus, all content on this wiki is available under the Creative Commons Attribution 4.0 license (or any later version). For futher information, please consult the official website of Creative Commons.
This website was designed with Bootstrap (4.1.3). Bootstrap is a front-end library of component for html, css and javascript. It relies on both Popper and jQuery. For further information, please consult the official website of Bootstrap.

