| Line 596: | Line 596: | ||
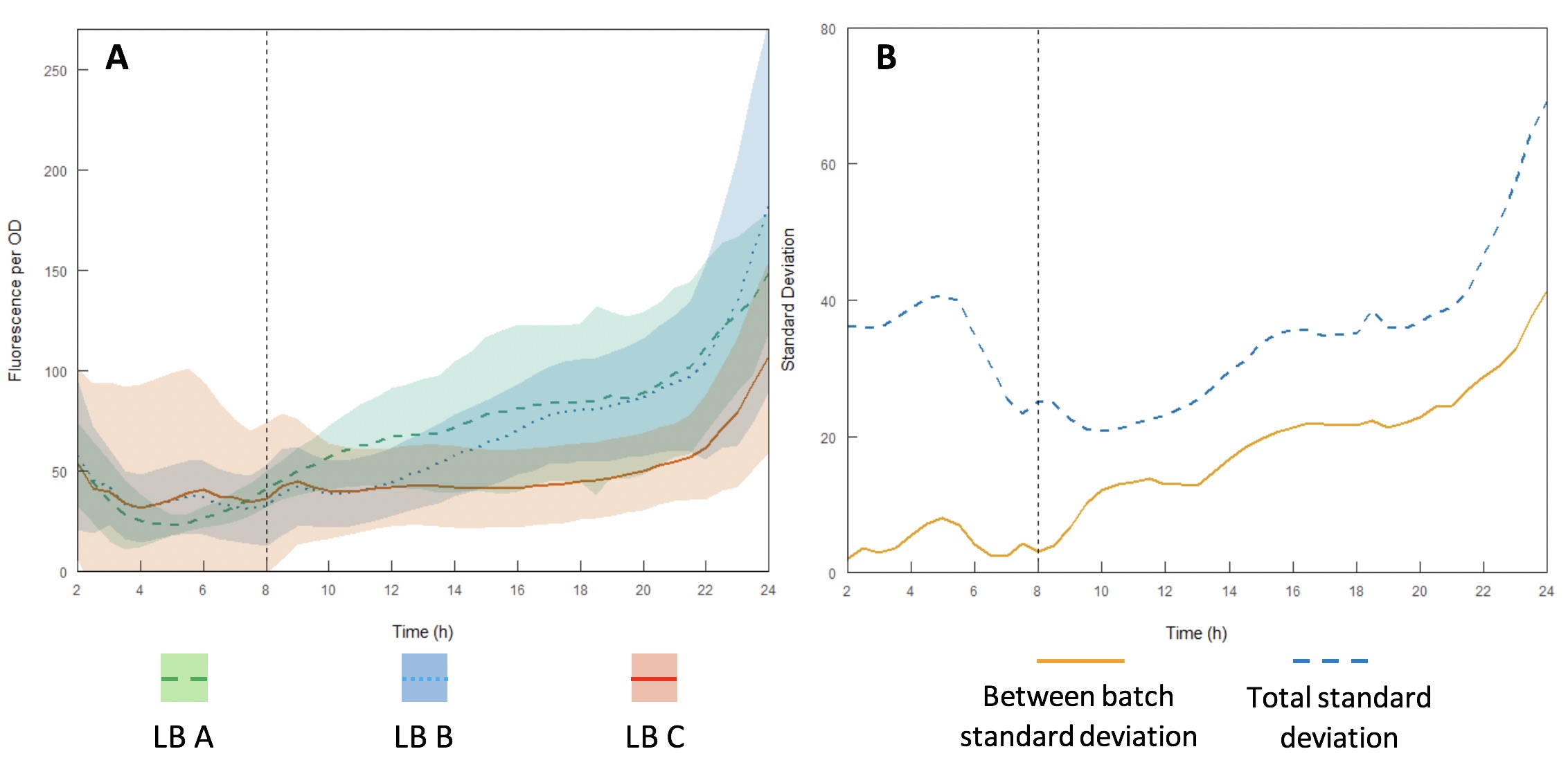
<p>Growth curves were performed using LB A-C (Figures 13A and B) to evaluate variability between batches and within each individual batch. Interestingly, the variation within each batch was greater than the variation between batches, suggesting that the differences in composition of yeast extract and tryptone can be accounted for by having a large enough sample size. However, for comparing expression between studies, total standard deviation is considerably higher. As such, differences in expression may be sufficient to affect the accuracy of comparisons of part performance across the literature.</p> | <p>Growth curves were performed using LB A-C (Figures 13A and B) to evaluate variability between batches and within each individual batch. Interestingly, the variation within each batch was greater than the variation between batches, suggesting that the differences in composition of yeast extract and tryptone can be accounted for by having a large enough sample size. However, for comparing expression between studies, total standard deviation is considerably higher. As such, differences in expression may be sufficient to affect the accuracy of comparisons of part performance across the literature.</p> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/2/2c/T--Newcastle--MeasurementFigure13.jpg"style="width: | + | <img src="https://static.igem.org/mediawiki/2018/2/2c/T--Newcastle--MeasurementFigure13.jpg"style="width:40%"> |
<p><center>Figure 13 – A: fluorescence per OD600 for recombinant E. coli DH5α expressing the iGEM interlab positive control device GFP gene E0040 in three batches (designated A, B and C) of LB broth made with tryptone and yeast extracts from different batches and different manufacturers with standard deviations (shaded areas); B: standard deviation of fluorescence per OD between each batch and total standard deviation across the whole dataset. Dotted vertical line represents the end of exponential growth phase and start of stationary phase.</p> </center> | <p><center>Figure 13 – A: fluorescence per OD600 for recombinant E. coli DH5α expressing the iGEM interlab positive control device GFP gene E0040 in three batches (designated A, B and C) of LB broth made with tryptone and yeast extracts from different batches and different manufacturers with standard deviations (shaded areas); B: standard deviation of fluorescence per OD between each batch and total standard deviation across the whole dataset. Dotted vertical line represents the end of exponential growth phase and start of stationary phase.</p> </center> | ||
| Line 620: | Line 620: | ||
<p> Development of a defined media in which variability may be reduced would benefit efforts to attain greater reproducibility and standardisation. Towards this end, we modelled how components of a defined media affected E. coli growth. Growth in defined rich media was assessed and the experimental execution was performed using the OT-2 pipetting robot (Opentrons) (python code for OT-2 protocol available in X). After 10 hours, growth was recorded in several runs, with growth in ten runs reaching stationary phase after 24 hours (Figure 14). </p> | <p> Development of a defined media in which variability may be reduced would benefit efforts to attain greater reproducibility and standardisation. Towards this end, we modelled how components of a defined media affected E. coli growth. Growth in defined rich media was assessed and the experimental execution was performed using the OT-2 pipetting robot (Opentrons) (python code for OT-2 protocol available in X). After 10 hours, growth was recorded in several runs, with growth in ten runs reaching stationary phase after 24 hours (Figure 14). </p> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/a/aa/T--Newcastle--MeasurementFigure14.jpg"style="width: | + | <img src="https://static.igem.org/mediawiki/2018/a/aa/T--Newcastle--MeasurementFigure14.jpg"style="width:40%"> |
<p><center>Figure 14 – Growth curves for <i>E. coli</i> DH5α in rich defined media DoE runs over 22 hours.</p></center> | <p><center>Figure 14 – Growth curves for <i>E. coli</i> DH5α in rich defined media DoE runs over 22 hours.</p></center> | ||
| Line 626: | Line 626: | ||
<p>The OD600 values at 24 hours were used to build a Projection to Latent Structures (or Partial Least Squares (PLS)) model using JMP software. This allowed visualisation of factor effects on E. coli growth (Figure 15). It is observed that there are positive effects on growth of increasing citrate, thiamine and tricine concentration, while there are negative effects of increasing K2HPO4. Additionally, four interactions between components in the media were shown to be important predictors of DH5α growth. These are interactions between citrate and tricine (VIP = 1.45), thiamine and EDTA (VIP = 1.27), thiamine and tricine (VIP = 1.20) and citrate and thiamine (VIP = 1.11). The media composition predicted by the model to maximise DH5α growth is shown in Figure 15.</p> | <p>The OD600 values at 24 hours were used to build a Projection to Latent Structures (or Partial Least Squares (PLS)) model using JMP software. This allowed visualisation of factor effects on E. coli growth (Figure 15). It is observed that there are positive effects on growth of increasing citrate, thiamine and tricine concentration, while there are negative effects of increasing K2HPO4. Additionally, four interactions between components in the media were shown to be important predictors of DH5α growth. These are interactions between citrate and tricine (VIP = 1.45), thiamine and EDTA (VIP = 1.27), thiamine and tricine (VIP = 1.20) and citrate and thiamine (VIP = 1.11). The media composition predicted by the model to maximise DH5α growth is shown in Figure 15.</p> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/d/d4/T--Newcastle--MeasurementFigure15.jpg"style="width: | + | <img src="https://static.igem.org/mediawiki/2018/d/d4/T--Newcastle--MeasurementFigure15.jpg"style="width:40%"> |
<p><center>Figure 15 – Prediction profiler for the model of factors affecting growth of Escherichia coli DH5α in a defined rich medium after 24 hours. The vertical red dashed line represents the factor concentration or level in the medium, changing which alters the horizontal red dashed line, representing culture OD600. Statistically significant factors highlighted in green were citrate, thiamine, tricine and KH2HPO4 (VIP = 2.39, VIP = 2.14 and VIP = 1.99, VIP = 1.04 respectively). Factor levels are set to max desirability, shown in red text beneath each x-axis (i.e. levels producing the maximum OD600). The model was constructed using JMP Pro v.12 statistical software (SAS Institute Inc.), based on experimental results of a definitive screening based experimental design.</p></center> | <p><center>Figure 15 – Prediction profiler for the model of factors affecting growth of Escherichia coli DH5α in a defined rich medium after 24 hours. The vertical red dashed line represents the factor concentration or level in the medium, changing which alters the horizontal red dashed line, representing culture OD600. Statistically significant factors highlighted in green were citrate, thiamine, tricine and KH2HPO4 (VIP = 2.39, VIP = 2.14 and VIP = 1.99, VIP = 1.04 respectively). Factor levels are set to max desirability, shown in red text beneath each x-axis (i.e. levels producing the maximum OD600). The model was constructed using JMP Pro v.12 statistical software (SAS Institute Inc.), based on experimental results of a definitive screening based experimental design.</p></center> | ||
Revision as of 11:28, 17 October 2018