Schematic diagram

BBa_K2601010

We combined Frb, yEGFP and HOTag6 togethor in BBa_K2601010. Frb can interact with FKBP after adding rapamycin, which makes the RapaSPOT formation controllable. We constructed yeast strains with FKBP-mCherry-HOTag3 and Frb-yEGFP-HOTag6 and used 10 μM rapamycin to induce SPOT formation. Minutes after adding rapamycin, granules appeared and became larger gradually.
BBa_K2601011

To make our SPOT a reaction hub, we fused the enzymes of β-carotene synthase system, CrtI, CrtE and CrtYB into Frb-HOTag6 backbone. In the presence of rapamycin, FKBP-yEGFP-HOTag3 and Frb-enzyme-HOTag6 can condense into aggregates. And if the yeast contains SPOT, there will be more β-carotene producted. Meanwhile, we wondered if we can load enzymes onto SPOT mediated by indirected connection. We fused CFP to anti-GFP nanobody as the demonstration of our design and we observed the co-localization of the blue and green fluorescence as expected.
BBa_K2601012

In the biobrick BBa_K2601012, we fused SUMO, yEGFP and HOTag3 together. SUMO is the interaction module which can bind SIM spontaneously, while HOTag3 can introduce multivalence. These two features are necessary for SPOT formation. We expressed SUMO-yEGFP-HOTag3 and SIM-mCherry-HOTag6 in one yeast strain, and we observed the colocalization of yEGFP and mCherry in two different fluorescence channels, which confirms our hypothesis that the two components could form synthetic organelles.
| BBa_ | Part |
| K2601000 | Promoter-tet07 |
| K2601001 | Promoter-PDH3 |
| K2601002 | SUMO |
| K2601003 | SIM |
| K2601004 | HOTag3 |
| K2601005 | HOTag6 |
| K2601007 | Frb-yEGFP |
| K2601008 | FKBP-yEGFP |
| K2601010 | Frb-yEGFP-HOTag6 |
| K2601011 | FKBP-yEGFP-HOTag3 |
| K2601012 | SUMO-yEGFP-HOtag3 |
| K2601021 | Tet07-Frb-yEGFP |
| K2601023 | PDH3-Frb-yEGFP |
| K2601025 | Tet07-FKBP-yEGFP |
| K2601026 | TEF1-FKBP-yEGFP |
| K2601027 | PDH3-FKBP-yEGFP |
| K2601032 | Tet07-Frb-yEGFP-HOTag6 |
| K2601033 | TEF1-Frb-yEGFP-HOTag6 |
| K2601034 | PDH3-Frb-yEGFP-HOTag6 |
| K2601037 | TEF1-FKBP-yEGFP-HOTag3 |
| K2601038 | PDH3-FKBP-yEGFP-HOTag3 |
| K2601040 | Tet07-SUMO-yEGFP-HOtag3 |
| K2601041 | TEF1-SUMO-yEGFP-HOtag3 |
| K2601042 | PDH3-SUMO-yEGFP-HOtag3 |
| K2601054 | PDH3-Frb-crtE-HOTag6 |
| K2601056 | Tet07-Frb-crtYB-HOTag6 |
| K2601058 | PDH3-Frb-crtYB-HOTag6 |
| K2601060 | SIM-crtE-Hotag6 |
| K2601061 | SIM-crtYB-Hotag6 |
BBa_K2601004
For this year’s project, we chose BBa_K2601004 which codes homo-oligomeric tag 3 (short as HOTag3) for the special award of the best Basic Part. HOTags are the biobricks that we used to introduce multivalence in our synthetic organelles. In natural process, phase separation occurred depending on multiple repeats protein domains, such as the interaction between Nephrin which three phosphotyrosine motifs and the SRC homology 2 (SH2) domain on Nck [1]. Using multiple repeat domains in a man-made design is not ideal, because it would not only make the scaffold extremely large but also be problematic for molecular cloning and making transgenic yeasts. Thus, instead of using multiple repeats, we turned to use the de novo-designed HOTags. These HOTags contain approximately 30 amino acids. HOTag3 has high stoichiometry, forming hexamer spontaneously[2].
We fused two sets of interaction module into the multivalence module. First, we fused SUMO to hexameric HOTag3 and SIM to tetrameric HOTag6. With the result of fluorescence imaging (Fig. 1), we confirmed that these two recombinant proteins can form granules successfully. Meanwhile we used FRAP to verify if the granule is liquid-like. After photobleaching, the fluorescence of granules quickly recovered, which indicated that the granules had rapid mass exchange with cytoplasm.

Figure. 1B The state of the synthetic organelle was observed by fluorescence recovery after photobleaching(FRAP). After photobleaching, fluorescence recovered in eight seconds. Scale bar, 1.70μm.
Furthermore, we used Tet07, a doxycycline-inducible promoter, to control the expression of the SIM component. SIM fused with HOTag6 couldn’t express in the absence of doxycycline. Before we added doxycycline, SUMO-HOTag3 was evenly distributed in the cells and can’t phase separate. But after we added it, phase separation gradually appears (Fig. 2).
 Figure. 2 Screenshots taken from the live-cell videos. Before adding dox, only green fluorescence could be detected. After adding dox, colocalization of red and green puncta could be captured. Scale bar, 30μm
Figure. 2 Screenshots taken from the live-cell videos. Before adding dox, only green fluorescence could be detected. After adding dox, colocalization of red and green puncta could be captured. Scale bar, 30μm
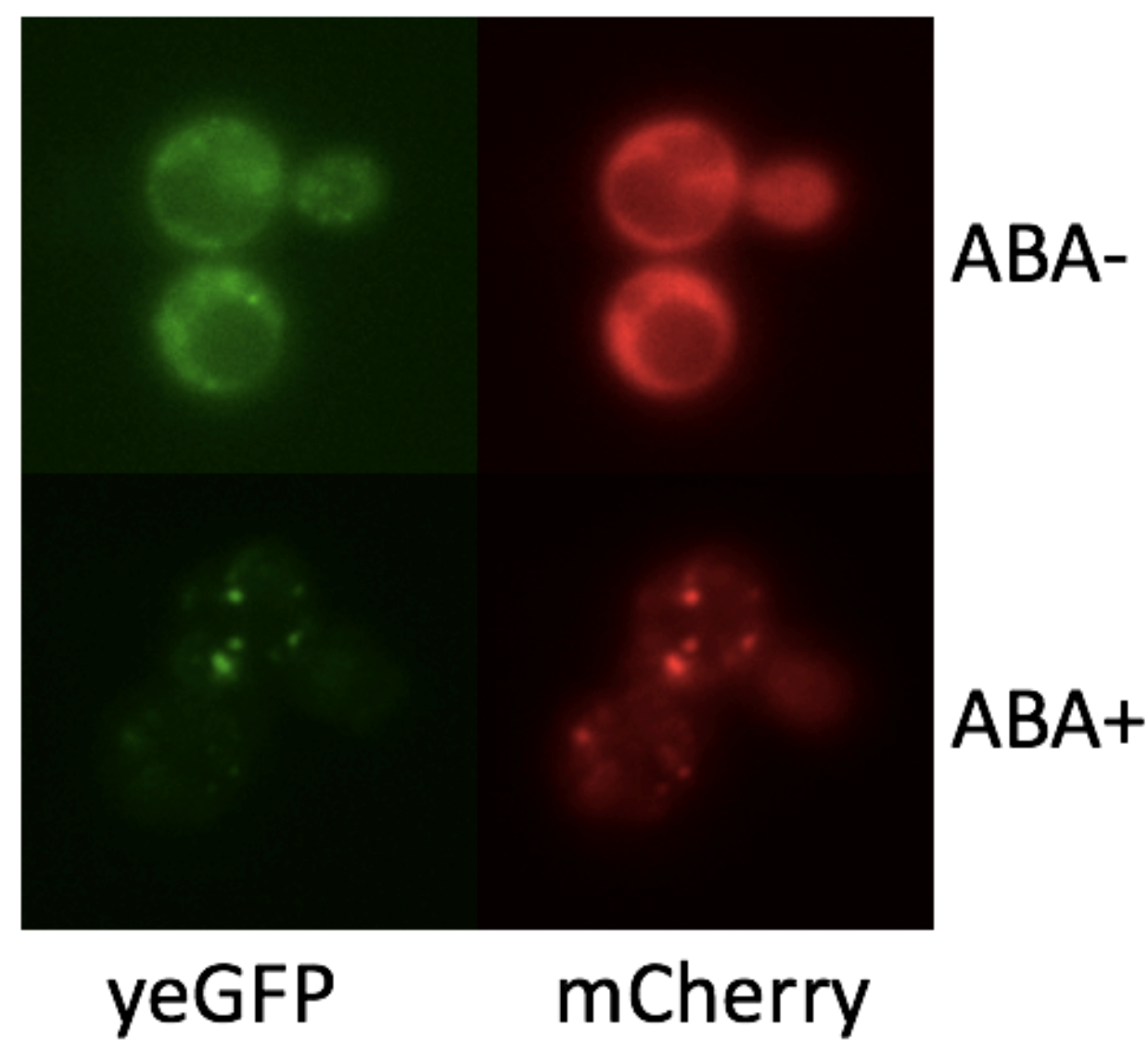
To demonstrate if we can control the formation of synthetic organelles at protein-level, we transformed the SUMO-SIM system to rapamycin-induced dimerization system (FKBP-Frb). Phase separation happens only if we add rapamycin to the culture. From the result of fluorescence imaging, we observed only the concentration of FKBP-mCherry-HOTag3 and Frb-yEGFP-HOTag6 is appropriate, they can combine with each other successfully (Fig. 3).

we can use the synthetic phase separation-based organelle platform as a sensor.
In a word, with the basic part BBa_K2601004, we can achieve the formation of the synthetic phase separation-based organelle platform in many different conditions. Based on this part, we designed a series of composite part.
[1] Banani, S. F., Lee, H. O., Hyman, A. A., & Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology, 18(5), 285.
[2] Woolfson D N, Bartlett G J, Burton A J, et al. (2015). De novo protein design: how do we expand into the universe of possible protein structures?. Current opinion in structural biology, 33: 16-26.
BBa_K2602011
We chose BBa_K2602011 which can express protein FKBP-yEGFP-HOTag3 to apply for the award of the Best Composite Part.
A widespread use of rapamycin has been developed to take advantage of the small molecule’s ability to heterodimerize proteins. Rapamycin binds to the FK506 binding protein (FKBP) as well as to a 100-amino acid domain of the mammalian target of rapamycin (mTOR), known as the FKBP-rapamycin binding domain (Frb). Proteins of interest can be expressed as fusions to FKBP or Frb, and then conditionally dimerized by adding rapamycin.
To make our granules controllable, we fused the interaction module FKBP, the reporter yEGFP and the multivalence module HOTag3 together. Combined with another recombinant protein Frb-HOTag6, this composite part can respond to rapamycin in the environment and form a synthetic phase separation-based organelle, which is confirmed by our result (Fig. 5).

To regulate the concentration of FKBP-HOTag3 and Frb-HOTag6, we found that only in an appropriate condition that only when FKBP-HoTag3 had a low level expression while Frb-HoTag6 had a high level expression, granules formed (Fig. 6). The experimental results was consistent with the model.

Rapamycin is a famous kind of antifungi reagent. We found FKBP-Frb based phase separation system could alleviate the inhibitory effect of rapamycin. It could sequester rapamycin in the granule and rescue the yeast from the toxicity of the rapamycin. From the yeast growth curves we could see that there was a significant difference between cells with and without phase separation.
 Figure. 7 The growth curve of yeasts with and without SPOT. The groups with SPOT show stronger resistance to rapamycin.
Figure. 7 The growth curve of yeasts with and without SPOT. The groups with SPOT show stronger resistance to rapamycin.
Based on this composite part, we demonstrate if we can introduce more functions into our synthetic organelles, such metabolic regulation. We fused three enzymes named CrtI, CrtYB, and CrtE which can produce β-carotene, into the Frb-HOTag6. We wondered if our synthetic organelles can increase the production rate. Results from photograph and HPLC confirmed our hypothesis (Fig. 8).

Figure. 8B Design of SPOT producing carotene.
Figure. 8C Fluorescence images and carotene production result after rendered by rapamycin for 36h. Figure. 8D HPLC result of carotene production after 48h.
2018 Peking iGEM team members devoted themselves to constructing a part collection that can drive phase separation. We have not only submitted all of new basic HOTag parts, but also provided multiple composite parts combining HOTags with other protein-protein interaction modules and fluorescent reporters. The basic part, HOTag3, is a homo-oligomeric short peptide containing only 30 amino acids. It has high stoichiometry, forming hexamer spontaneously. The HOTag3, together with another tetrameric HOTag6, can robustly drive phase separation upon protein-protein interaction. Protein-protein interaction is achieved by our dimerization parts, including FKBP/Frb and SUMO/SIM. Functions of all the parts were thoroughly tested. Some thermodynamic and kinetic properties of the parts were characterized as well. We believe the HOTag is useful tool for other iGEM teams to investigate protein phase separation and design synthetic organelles.

