Design |
Bacterial Evolutionary Game Simulation (BEGS)
for Snowdrift, Harmony, Stag Hunt and Prisoner's Dilemma Games

- The mechanism is as follows.
- The producer (cooperator) constitutively expresses beta-glucosidase on its surface and degrades the cellobiose.
- The cooperator and the cheater take them as energy source and the cheater expresses GFPuv as a reporter gene when it takes up the glucose.
- As the ratio between the cheater and cooperator changes, the total number of cooperator and cheater changes at the end.
- Constitutive expression vector for both cheater and cooperator
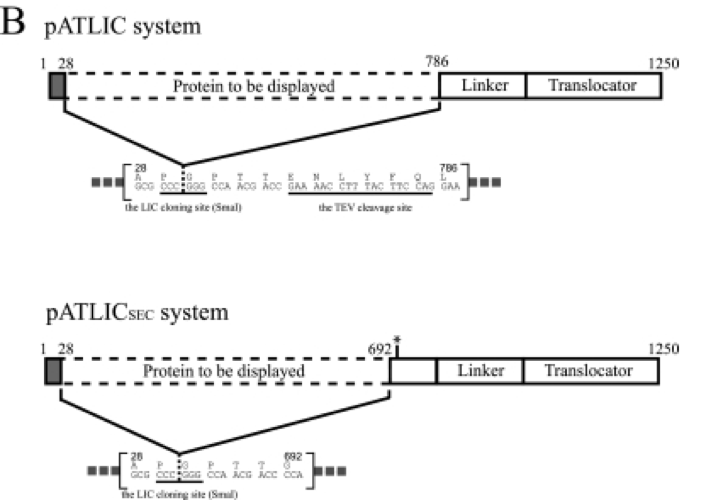
- Using the plasmid containing BBa_J23106 showing high-level constitutive expression, the constitutive expression vector applicable to ligation independent cloning including SwaI restriction site at the LIC site was constructed. Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. The resultant LIC vector was designated as ‘pCELIC’.
- Bgl1A of S. degradans was subcloned into a pATLIC vector as a previously report for the autodisplay (Ko et al., 2012) and was amplified by LA-taq polymerase by PCR. GFPuv also was amplified using α-taq polymerase by PCR. Amplified products were mixed with the linear LIC ready pCELIC vector at 1 1:4 molar ratios before transforming into DH5α.
- BBa_J23106 Part-only sequence (35 bp)
- tttacggctagctcagtcctaggtatagtgctagc
[Figure 1. pATLIC display system for the designed vector]
- E.coli BW25113 as an expression host
- As E.coli BW25113 is used as an display and expression host. In our experiment, cooperator expresses beta-glucosidase on its cell surface and degrades cellobiose into glucose. It is done by a surface display system using the autotransporter YfaL protein.
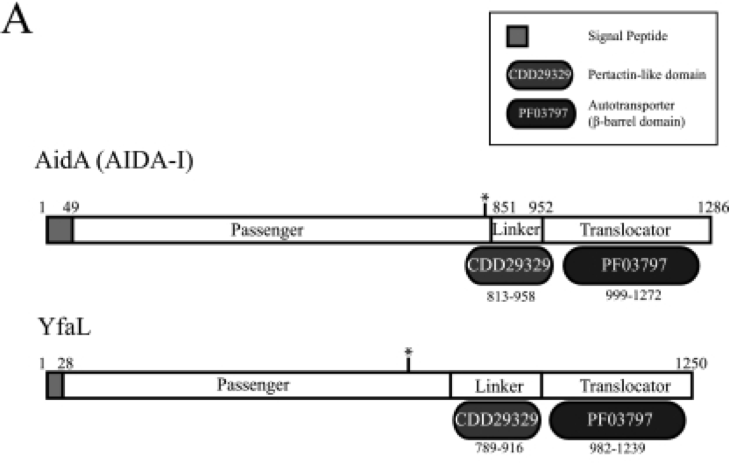
[Figure 2. Schematic diagrams for the domain organization of autotransporter]
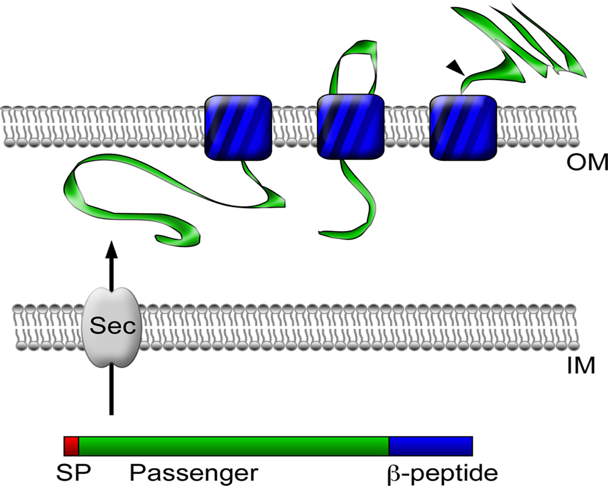
[Figure 3. Autotransporter protein structure]
References
- Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., . . . Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Molecular Systems Biology, 2. doi:10.1038/msb4100050
- Ko, H., Park, E., Song, J., Yang, T. H., Lee, H. J., Kim, K. H., & Choi, I. (2012). Functional Cell Surface Display and Controlled Secretion of Diverse Agarolytic Enzymes by Escherichia coli with a Novel Ligation-Independent Cloning Vector Based on the Autotransporter YfaL. Applied and Environmental Microbiology, 78(9), 3051-3058. doi:10.1128/aem.07004-11
- https://www.researchgate.net/figure/Autotransporter-protein-structure-and-secretion-Autotransporter-proteins-have-modular_fig1_51233167
- As E.coli BW25113 is used as an display and expression host. In our experiment, cooperator expresses beta-glucosidase on its cell surface and degrades cellobiose into glucose. It is done by a surface display system using the autotransporter YfaL protein.
Mechanisms
Sponsors







