Construct design
To characterize our system, we needed to make various plasmid constructs containing various combinations of suitable reporter proteins with and without secretion signals, unfolding chaperones and additional translocon protein.
These constructs will be used in combination with the Synthetic Injector E. coli (SIEC) strain, which have the 5 operons encoding the full injectisome chromosomally integrated under IPTG inducible promoters [1]. The constructs will allow us to characterize any combination of genes by combining up to two vectors with both arabinose and IPTG induction.
1. Components
1.1 Reporter proteins
Good reporters for proof of concept should be easily expressed, detectable and without post-translational modifications. We chose to work with fluorescent reporters, sfGFP, mCherry and a small luciferase enzyme, NanoLuc. This will allow us to easily characterize our system by measuring both expression and injection of protein through membranes by fluorescent measurement and fluorescent microscopy. We chose BBa_I746916 (sfGFP) and BBa_J06504 (mCherry) from the 2018 iGEM distribution kit and received BBa_K1159001 (NanoLuc) upon request from iGEM HQ.
In Charpentier and Oswald 2004 [2], β-lactamase was used as a substrate for secretion by the E. coli injectisome. β-lactamase is thus a proven substrate, but experimentally it is harder to measure its activity than the fluorescent reporters. So we decided to use beta-lactamase (BBa_K1189008) from the 2018 iGEM distribution kit as one of our target protein.
Finally, we used only mCherry, sfGFP, β-lactamase for our experiments.
1.2 Map20 secretion signal
E. coli injectisome system naturally secrete many effector proteins into its host cell. According to Charpentier and Oswald, 2004 [2], 20 amino acid sequence at the N-terminal domain of four T3SS effector proteins, Map, Cif, Tir, and EspF, is an essential and sufficient signal for E. coli injectisome transport.
We chose to use 20 amino acid sequence from N-terminal part of Map protein as a T3SS secretion signal in our system, as it was demonstrated in Charpentier and Oswald, 2004 [2] to be the most efficient signal. We named it Map20. In our constructs we fused this signal N-terminally to the various reporter proteins.
1.3 Chaperones
For the Map20-tagged proteins to go through the injectisome, they will need to be in unfolded states. Charpentier and Oswald [2] showed that secretion of larger proteins through injectisomes from E. coli improves when chaperones are present. Therefore we chose to express the two E. coli chaperones CesT and CesF. These chaperones assist natural effector protein transport and are not present in the modified LEE region in SIEC strain.
1.4 Translocon protein
EspD is a subunit of the injectisome which can be inserted into the host membrane. We plan to use it as a docking site to facilitate injectisome binding to the membrane. This creates a docking site, and will be useful in the case that the bacteria can not recognize and dock to the membrane we use. EspD has previously been used to create a pore on a lipid membrane (Chatterjee et al. 2015 [3]). EspD is already expressed in the SIEC strain, but it could be important to express more for higher binding efficiency to membranes.
2. Expression vectors with reporter proteins
We based our expression vectors on the BBa_K228005 biobrick. In this vector, expression of a reporter is thus controlled by an arabinose-inducible promoter (pBAD). The plasmid also contains a gene encoding the arabinose-responsive regulatory protein araC under control of a pCat promoter and an chloramphenicol resistance cassette.
From BBa_K228005 we constructed two different backbones, either with or without the genes for the 6x-His tagged effector chaperones CesT and CesF, see figure 1.A and figure 1.B. The chaperone genes are positioned between the araC gene and the downstream terminator and are thus controlled by the constitutive promoter pCat.
These backbones are combined with a reporter protein tagged or untagged with Map20 signal sequence, see figure 1.C and figure 1.D. The reporter proteins are in both cases 6x-His tagged and controlled by pBAD. The Map20 is linked to the reporter protein by a glycine-serine linker peptide.

Figure 1. A vector backbone is constructed based on BBa_K228005 either without (A) or with chaperones CesT and CesF, shown in dark green on figure, (B). A reporter gene (sfGFP, mCherry, NanoLuc or β-lactamase) is inserted downstream of the pBAD promoter. The reporter gene is either untagged (C) or tagged with Map20 (D), shown in red, at its N-terminal. The figure was made using SnapGene.
This way we have four different expression plasmids pr reporter protein. In total we constructed 14 different plasmids with reporter proteins. An overview of the constructs can be seen below.
| Plasmid constructed | Reporter | N-terminal Map20 | CesT and CesF |
| + | sfGFP | + | + |
| + | sfGFP | + | - |
| + | sfGFP | - | + |
| + | sfGFP | - | - |
| + | mCherry | + | + |
| + | mCherry | + | - |
| + | mCherry | - | + |
| + | mCherry | - | - |
| + | nanoluc | + | + |
| + | nanoluc | + | - |
| - | nanoluc | - | - |
| - | nanoluc | - | + |
| + | β-lactamase | + | + |
| + | β-lactamase | + | - |
| + | β-lactamase | - | - |
| + | β-lactamase | - | + |

Figure 2 : Example of constructed plasmid with mCherry reporter protein
3. Expression vector with the translocon protein EspD
For expression of the translocon protein EspD, we used BBa_K731500 as an expression vector. The EspD is inserted downstream of pTac promoter and can be induced.
The plasmid, seen on figure 3, encodes 6His-tagged EspD translocon under pTac, which can be induced by IPTG. The EspD translocon protein self-assembles into a hydrophobic membrane protein complex that is sufficient for membrane penetration and pore formation. This translocon protein is normally secreted along with the EspB hydrophilic translocon to penetrate the host cell membrane and serve as a docking site for the injectisome. This 6His-tagged EspD can be purified by Ni-NTA column or TALON column. It might be used to pre-assemble a translocon on a target membrane to induce injectisome docking on an artificial membrane.

Figure 3. EspD Translocon expression plasmid. The plasmid encodes the translocon protein EspD (shown in purple on the figure) in a BBa_K731500 backbone. The figure was made using SnapGene.
4. Experimental plan
The flow charts below show the experimental plan that was the basis of our work over the summer. Read more about the cloning strategy that we used when assembling our constructs here.
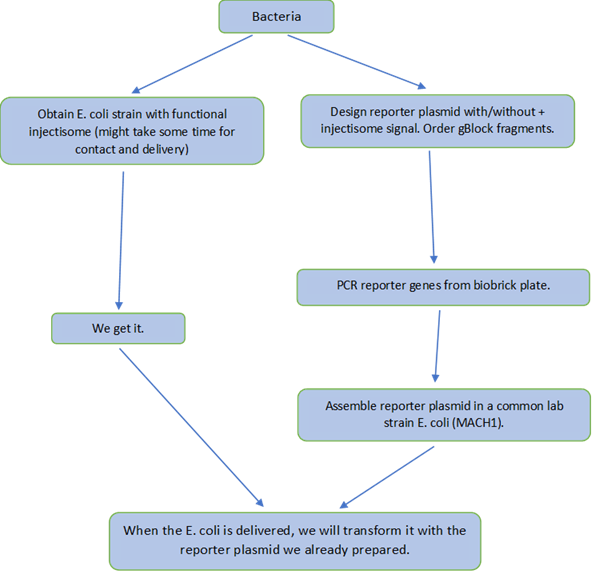
Figure 4. Flowchart describing the laboratory work concerning constructing expression vectors and finally transforming the SIEC strains with the plasmids.

Figure 5. Flow chart describing the different steps in designing and characterizing a home made chamber for protein production. We use an egg yolk membrane and test for leakiness of the system.

Figure 6. Flow chart describing the basis of our experiments combining membranes and SIEC bacteria. Initially we had planned to work with liposomes and an artificial membrane, which was later replaced with experiments with egg yolk membrane and plant protoplasts.

Figure 7. Flow chart describing experimental troubleshooting plan for secretion signal.
References
[1] Ruano-Gallego, D., Álvarez, B., Fernández, L. A. (2015) Engineering the Controlled Assembly of FIlamentous Injectisomes in E. Coli K-12 for Protein Translocation into Mammalian Cells. ACS Synth. Biol. 4, pp 1030-1041.
[2] Charpentier, X., Oswald, E. (2004) Identification of the Secretion and Translocation Domain of the Enteropathogenic and Enterohemorrhagic Escherichia coli Effector Cif, Using TEM-1 β-Lactamase as a New Fluorescence-Based Reporter. J Bacteriol, 186(16), pp 5486-5495.
[3] Chatterjee, A., Caballero-Franco, C., Bakker, D., Totten, S., Jardim, A. (2015) Pore-forming Activity of the Escherichia coli Type III Secretion System Protein EspD. J Biol Chem. 290 (42) pp. 25579-25594.


