Collaborations
Pasteur Paris
This year, we, the WPI iGEM team, collaborated with the Pasteur Paris iGEM Team. Both of our teams conducted research on biofilms so we decided to test each other’s protocols for quantifying biofilm growth to make sure the data was consistent throughout different labs. This comparison will be used to prove that any data collected using these techniques is accurate because of the reproducibility. If the data between the labs is significantly variable, then we know that the data collected depends heavily on how exactly the protocol was carried out, so the protocol should be edited to make it more consistent.
Our data concluded that the plate rinsing protocol that we followed was more effective than that of the Pasteur Paris iGEM team because submerging the plate in water is more likely to cause contamination, even into empty wells. We successfully grew biofilms using this protocol, but the data we collected shows the most biofilm in the empty column, which must be a product of contamination during the rinsing process. If we had more time, we would work further with Pasteur Paris to perfect a method of quantifying biofilms.
Pasteur Paris Collaborations Page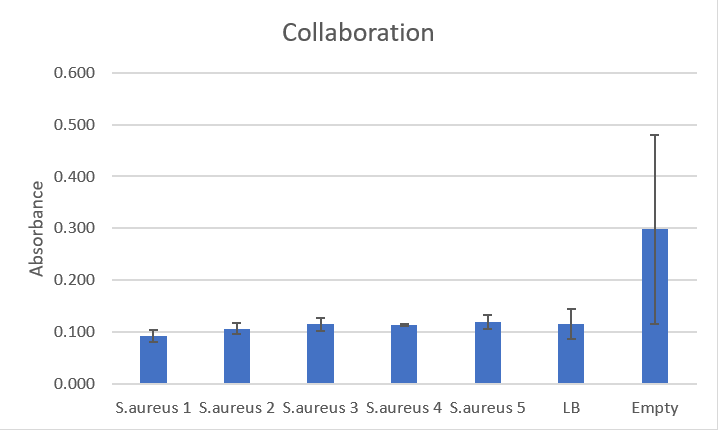
Graph
This graph shows the massive amount of error associated with this method of rinsing plates, especially in the empty wells that should not have had any biofilm at all.
Massachusetts Institute of Technology (MIT)
We also collaborated with the MIT iGEM team. We held a skype call to discuss different means of quantifying biofilms and we provided them with our crystal violet biofilm assay protocol, and they shared their colony forming units of planktonic cells protocol with us. We used this protocol when testing biofilm formation on lettuce leaves.
MIT Collaborations Page


