| Line 3: | Line 3: | ||
<meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | ||
<title>Nanjing-China2018</title> | <title>Nanjing-China2018</title> | ||
| − | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Nanjing-China/CSS: | + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Nanjing-China/CSS:4?action=raw&ctype=text/css" /> |
<link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Nanjing-China/CSS:loader?action=raw&ctype=text/css" /> | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Nanjing-China/CSS:loader?action=raw&ctype=text/css" /> | ||
<style type="text/css"> | <style type="text/css"> | ||
| − | |||
#HQ_page .word-note p{ text-align:center; } | #HQ_page .word-note p{ text-align:center; } | ||
| − | |||
</style> | </style> | ||
<script type="text/javascript" src="https://2018.igem.org/Team:Nanjing-China/Javascript:1?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2018.igem.org/Team:Nanjing-China/Javascript:1?action=raw&ctype=text/javascript"></script> | ||
| Line 71: | Line 69: | ||
$("#loader").fadeOut("slow"); | $("#loader").fadeOut("slow"); | ||
}) | }) | ||
| − | |||
</script> | </script> | ||
<div id="d"> | <div id="d"> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/f/fe/T--Nanjing-China--signal-1.jpg" width="100% | + | <img src="https://static.igem.org/mediawiki/2018/f/fe/T--Nanjing-China--signal-1.jpg" width="100%" onclick="MM_effectBlind('HOME', 1000, '0%', '100%', true)" /> |
<div align="center"> | <div align="center"> | ||
<div id="HOME"> | <div id="HOME"> | ||
| − | + | <div class="sub"> | |
| − | <ul><li><a href="https://2018.igem.org/Team:Nanjing-China/ | + | <ul><li><a href="https://2018.igem.org/Team:Nanjing-China/Results">Results</a></ul></li></div> |
| − | + | <ul> | |
| − | + | <li><a href="#cds">CdS</a></li> | |
| − | + | <li><a href="#nit"><font size="-1">Nitrogen fixation</font></a></li> | |
| − | + | </ul> | |
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 93: | Line 88: | ||
<li><a href="https://2018.igem.org/Team:Nanjing-China">PEOPLE</a> | <li><a href="https://2018.igem.org/Team:Nanjing-China">PEOPLE</a> | ||
<ul> | <ul> | ||
| − | <li><a href="https://2018.igem.org/Team:Nanjing-China/Team"> | + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Team">TEAM</a></li> |
<li><a href="https://2018.igem.org/Team:Nanjing-China/Members">Members</a></li> | <li><a href="https://2018.igem.org/Team:Nanjing-China/Members">Members</a></li> | ||
<li><a href="https://2018.igem.org/Team:Nanjing-China/Attributions">Attributions</a></li> | <li><a href="https://2018.igem.org/Team:Nanjing-China/Attributions">Attributions</a></li> | ||
| Line 114: | Line 109: | ||
<li><a href="https://2018.igem.org/Team:Nanjing-China/Composite_Part">Composite</a></li> | <li><a href="https://2018.igem.org/Team:Nanjing-China/Composite_Part">Composite</a></li> | ||
<li><a href="https://2018.igem.org/Team:Nanjing-China/Improve">Improve</a></li> | <li><a href="https://2018.igem.org/Team:Nanjing-China/Improve">Improve</a></li> | ||
| − | + | </ul> | |
| − | + | </li> | |
| − | <li><a href="https://2018.igem.org/Team:Nanjing-China/Model">MODEL</a> | + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Model">MODEL</a></a> |
</li> | </li> | ||
<li><a href="https://2018.igem.org/Team:Nanjing-China/Human_Practices">PRACTICES</a> | <li><a href="https://2018.igem.org/Team:Nanjing-China/Human_Practices">PRACTICES</a> | ||
| Line 127: | Line 122: | ||
<li><a href="https://2018.igem.org/Team:Nanjing-China/Notebook">NOTEBOOK</a></li> | <li><a href="https://2018.igem.org/Team:Nanjing-China/Notebook">NOTEBOOK</a></li> | ||
</ul> | </ul> | ||
| − | + | </div> | |
| − | <div class="header"><img src="https://static.igem.org/mediawiki/2018/4/40/T--Nanjing-China--title-4.png" width="100%" onload="MM_effectAppearFade(this, 1000, 0, 100, false);MM_effectBlind('HOME', 1000, '0%', '100%', true)"></div> | + | <div class="header"><img src="https://static.igem.org/mediawiki/2018/4/40/T--Nanjing-China--title-4.png" width="100%" onload="MM_effectAppearFade(this, 1000, 0, 100, false);MM_effectBlind('HOME', 1000, '0%', '100%', true)"> |
| − | + | </div> | |
| + | <div class="contain"> | ||
<div class="word"> | <div class="word"> | ||
<div style="position:absolute; top:-90px; z-index:3; left:-10px;"> | <div style="position:absolute; top:-90px; z-index:3; left:-10px;"> | ||
| − | <img src="https://static.igem.org/mediawiki/2018/ | + | <img src="https://static.igem.org/mediawiki/2018/d/da/T--Nanjing-China--PROJECT-r.jpg" width="50%" /></div> |
| + | <div class="word-1" style="height:20px;"></div> | ||
</div> | </div> | ||
| − | + | <div class="word" id="cds"> | |
| − | + | <h2>Biosynthesis of CdS semiconductor</h2> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/a/a7/T--Nanjing-China--TEX-EDX.jpg" width="100%" /> | |
| − | + | <p>TEM-EDX analysis of CdS semiconductor. a) TEM images of biosynthesized CdS semiconductor on the surface of an engineered E. coli cell. b) Elemental analysis using EDX system, the result show that the semiconductor on cell surface is mainly composed of cadmium and sulfide.</p> | |
| − | + | <p> </p> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/e/e1/T--Nanjing-China--toxicity-1.jpg" width="70%" /> | |
| − | + | <div class="word-background-block" style="height:10px;"></div> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/3/3e/T--Nanjing-China--toxicity-2.jpg" width="70%" /> | |
| + | <p>Toxicity test was conducted to determine the maximum amount of Cd<sup>2+</sup> that is agreeable for E. coli growth. Compared with the control group that doesn’t contain surface-display gene, our constructed E. coli strain is more sensitive to Cd<sup>2+</sup>, and its growth will be restricted When the Cd<sup>2+</sup> concentration is above 150μM. So we select 100μM as the final Cd<sup>2+</sup> concentration for our further assays.</p> | ||
| + | <p> </p> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/0/05/T--Nanjing-China--ICP-MS.jpg" width="70%" /> | ||
| + | <p>The amount of biosynthesized CdS semiconductor on the E. coli cell surface was measured using inductively coupled plasma mass spectrometry (ICP-MS). These data confirmed the surface-displayed PbrR-mediated biological precipitation of CdS semiconductor on the outer membranes of the cells.</p> | ||
| + | <p> </p> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/a/a9/T--Nanjing-China--UV.jpg" width="70%" /> | ||
| + | <p>We performed ultraviolet-visible (UV-vis) spectral measurements to directly determine the optical band gap energy of these CdS semiconductor and the photocatalytic capability for the biological precipitation of CdS semiconductor on the outer membranes of the bacterial cells. The lowest-energy transition of the biosynthesized CdS nanoparticles was detected in the visible region of the solar spectrum (Eg = 2.92 eV, labsorption = 424 nm), confirming the photocatalytic ability of the in situ biosynthesized CdS semiconductor.</p> | ||
| + | <p> </p> | ||
</div> | </div> | ||
| − | + | <div class="word" id="nit"> | |
| − | + | <h2>Light-driven nitrogen fixation in E. coli cells</h2> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/c/cf/T--Nanjing-China--QPCR1.jpg" width="40%" /> | |
| − | + | <div class="word-background-block" style="width:20px;"></div> | |
| − | + | <img src="https://static.igem.org/mediawiki/2018/d/dc/T--Nanjing-China--QPCR2.jpg" width="40%" /> | |
| − | + | <p>To verify the expression of nitrogenase gene, we conducted Real-time Quantitative PCR(QPCR) to detect the transcription level of nif gene cluster in engineered E. coli, using 16S DNA as an internal reference. The result provided the relative expression level of each nif gene in our constructed E. coli strain. After comparing the result with the ideal expression ratio in Paenibacillus CR1 and model the transcription, we plan to optimize the nif gene cluster by adding promoters or altering the position of genes.<br/> | |
| − | + | Nitrogenase can not only reduce dinitrogen to ammonia but also reduce ethylene to acetylene. Therefore, we use gas chromatography to detect the amount of acetylene reduced, and indirectly detect its nitrogen fixation activity. On the basis of these results, NH<sub>3</sub> production by our engineered E. coli cell–CdS hybrid system is directly related to the biosynthesized CdS semiconductors as well as illumination and anaerobic conditions.</p> | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<div class="footer"> | <div class="footer"> | ||
<div align="center"> | <div align="center"> | ||
| Line 206: | Line 189: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | </div> | |
<div class="f-b"><img src="https://static.igem.org/mediawiki/2018/5/58/T--Nanjing-China--footer-4.png" width="100%" /></div> | <div class="f-b"><img src="https://static.igem.org/mediawiki/2018/5/58/T--Nanjing-China--footer-4.png" width="100%" /></div> | ||
</div> | </div> | ||
Revision as of 11:43, 11 October 2018


Biosynthesis of CdS semiconductor
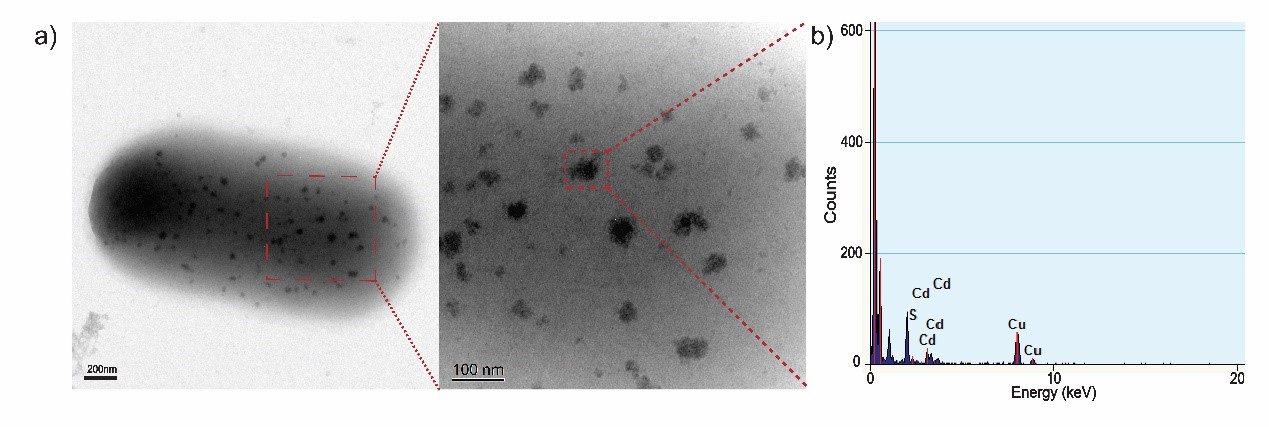
TEM-EDX analysis of CdS semiconductor. a) TEM images of biosynthesized CdS semiconductor on the surface of an engineered E. coli cell. b) Elemental analysis using EDX system, the result show that the semiconductor on cell surface is mainly composed of cadmium and sulfide.
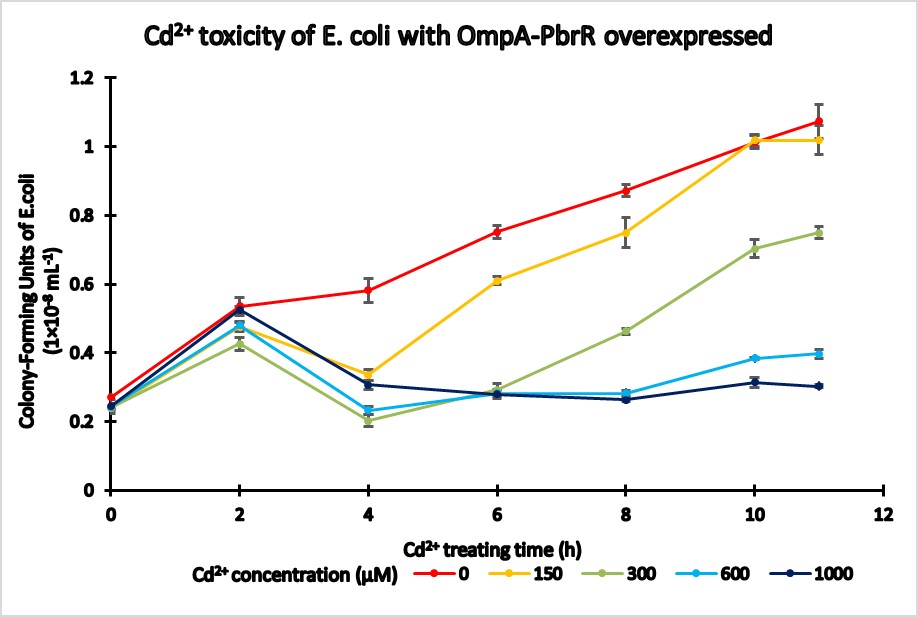

Toxicity test was conducted to determine the maximum amount of Cd2+ that is agreeable for E. coli growth. Compared with the control group that doesn’t contain surface-display gene, our constructed E. coli strain is more sensitive to Cd2+, and its growth will be restricted When the Cd2+ concentration is above 150μM. So we select 100μM as the final Cd2+ concentration for our further assays.

The amount of biosynthesized CdS semiconductor on the E. coli cell surface was measured using inductively coupled plasma mass spectrometry (ICP-MS). These data confirmed the surface-displayed PbrR-mediated biological precipitation of CdS semiconductor on the outer membranes of the cells.

We performed ultraviolet-visible (UV-vis) spectral measurements to directly determine the optical band gap energy of these CdS semiconductor and the photocatalytic capability for the biological precipitation of CdS semiconductor on the outer membranes of the bacterial cells. The lowest-energy transition of the biosynthesized CdS nanoparticles was detected in the visible region of the solar spectrum (Eg = 2.92 eV, labsorption = 424 nm), confirming the photocatalytic ability of the in situ biosynthesized CdS semiconductor.
Light-driven nitrogen fixation in E. coli cells

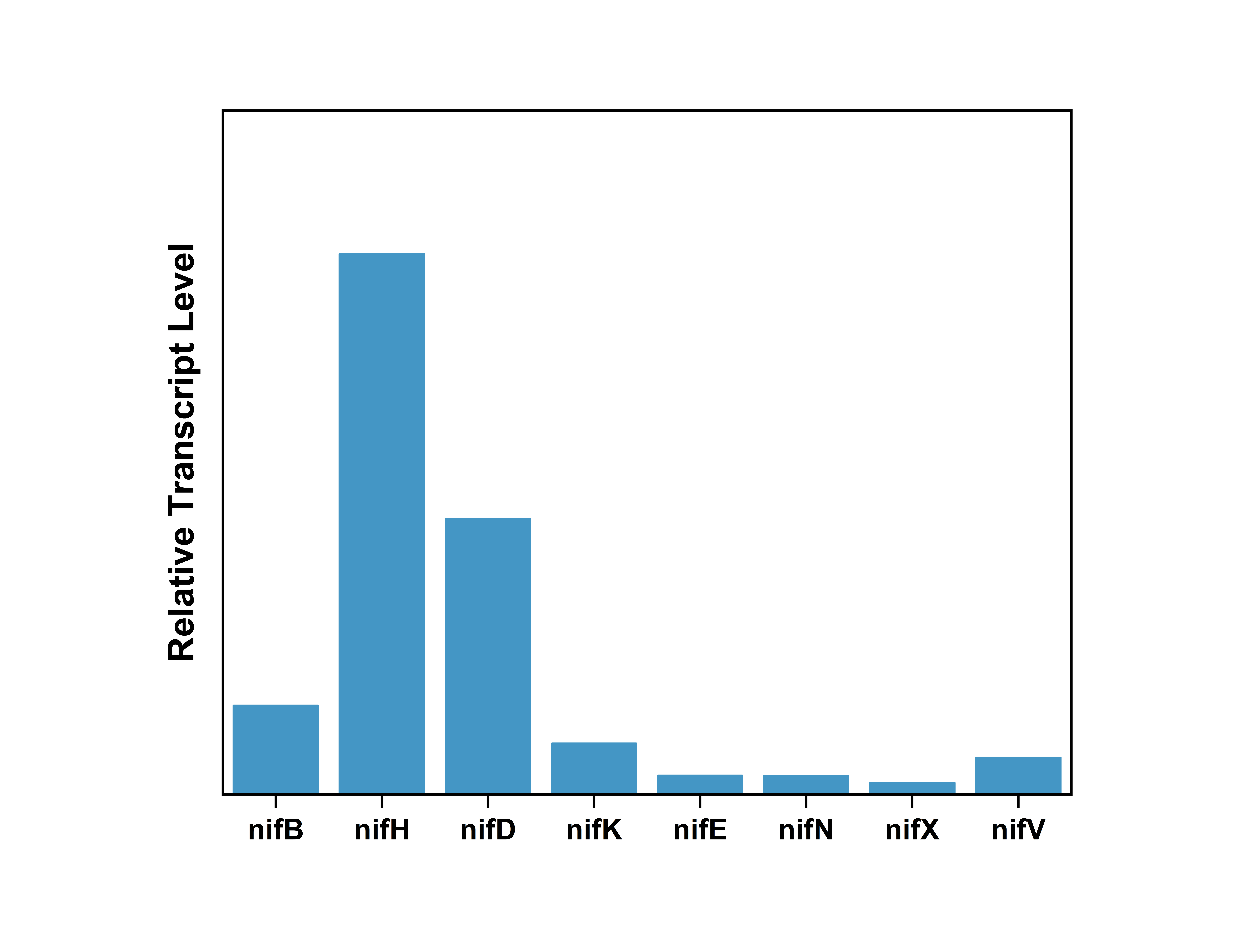
To verify the expression of nitrogenase gene, we conducted Real-time Quantitative PCR(QPCR) to detect the transcription level of nif gene cluster in engineered E. coli, using 16S DNA as an internal reference. The result provided the relative expression level of each nif gene in our constructed E. coli strain. After comparing the result with the ideal expression ratio in Paenibacillus CR1 and model the transcription, we plan to optimize the nif gene cluster by adding promoters or altering the position of genes.
Nitrogenase can not only reduce dinitrogen to ammonia but also reduce ethylene to acetylene. Therefore, we use gas chromatography to detect the amount of acetylene reduced, and indirectly detect its nitrogen fixation activity. On the basis of these results, NH3 production by our engineered E. coli cell–CdS hybrid system is directly related to the biosynthesized CdS semiconductors as well as illumination and anaerobic conditions.






