(Prototype team page) |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | {{SZU-China}} | + | {{SZU-China/header}} |
<html> | <html> | ||
| + | <style> | ||
| + | h1, h2 { | ||
| + | color: #469789; | ||
| + | } | ||
| + | </style> | ||
| + | |||
| + | <body data-spy="scroll" data-target="#myScrollspy" data-offset="10"> | ||
| + | <div class="container-full"> | ||
| + | <div class="card"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/6/67/T--SZU-China--interlab.jpg" /> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="container-fluid"> | ||
| + | <div class="row"> | ||
| + | <div class="col-9 offset-1 shadow"> | ||
| + | <div class="bs-component"> | ||
| + | <div class="jumbotron"> | ||
| + | <div class="container"></div> | ||
| + | <div class="center-block"> | ||
| + | <h1 id="header" class="text-center">InterLab Study</h1> | ||
| + | <p class="text-center" style="color:#469789 ;">"Measurement is fundamentally an act of communication." - Jacob Beal</p> | ||
| + | </div> | ||
| + | <p class="text-muted text-center">Reliable and repeatable measurement is a key component to all engineering disciplines. The same holds true for synthetic biology, which has also been called engineering biology. However, the ability to repeat measurements in different labs has been difficult. The Measurement Committee, through the InterLab study, has been developing a robust measurement procedure for green fluorescent protein (GFP) over the last several years. We chose GFP as the measurement marker for this study since it's one of the most used markers in synthetic biology and, as a result, most laboratories are equipped to measure this protein. | ||
| − | < | + | </p> |
| − | < | + | <center> |
| − | + | <a href="https://2018.igem.org/Measurement/InterLab" style="color:#469789 ;">More</a> | |
| − | < | + | </center> |
| − | </div> | + | </div> |
| + | </div> | ||
| + | <div class="indent"> | ||
| + | <h2 id="Calibration">Calibration</h2> | ||
| + | <h3>OD600 Reference point</h3> | ||
| + | <p>Measure the Abs600 of both the LUDOX CL-X (45% colloidal silica suspension) and water to obtain the correction factor. </p> | ||
| + | <div class="offset-1"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/6/6d/T--SZU-China--InterLab_OD600.png" width="360px" /> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="indent"> | ||
| + | <h3>Particle Standard Curve</h3> | ||
| + | <p>Measure the Abs600 of a series of dilution monodisperse silica microspheres to draw the Particle Standard Curve.</p> | ||
| − | <div class=" | + | <div class="row justify-content-center"> |
| + | <div class="col-8"> | ||
| − | <div class=" | + | <div class="card border-success mb-3"> |
| − | < | + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/d/d0/T--SZU-China--InterLab_FSC.png" alt="card-img-cap" /> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | </div> |
| − | < | + | <!-- |
| + | <div class="card border-success mb-3"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/4/48/T--SZU-China--InterLab_FSC_log.png" alt="card-img-cap" /> | ||
| + | |||
| + | </div> | ||
| + | --> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="indent"> | ||
| + | <h3>Fluorescence standard curve</h3> | ||
| + | <p>Measure the fluorescence of a series of diluted fluorescein to draw the fluorescein standard curve.</p> | ||
| + | <div class="row justify-content-center"> | ||
| + | <div class="col-8"> | ||
| + | <div class="card border-success mb-3"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/1/1b/T--SZU-China--InterLab_PSC.png" alt="card-img-cap" /> | ||
| + | |||
| + | </div> | ||
| + | <!-- | ||
| + | <div class="card border-success mb-3"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/6/6f/T--SZU-China--InterLab_PSC_log.png" alt="card-img-cap" /> | ||
| + | |||
| + | </div> | ||
| + | --> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="indent"> | ||
| + | <h2 class="">Cell measurement</h2> | ||
| + | <p> | ||
| + | We collaborate with the team SIAT-SCIE, because thire Distribution Kit was not delivered on time and we were during our final exam.<br /> So we provide our kit and get the streaks of the transformed devices from them. | ||
| + | </p> | ||
| + | <div class="row justify-content-center"> | ||
| + | <div class="col-8"> | ||
| + | <!-- | ||
| + | <div class="card border-success mb-3"> | ||
| + | <img width="512px" src="https://static.igem.org/mediawiki/2018/6/68/T--SZU-China--InterLab_Abs.jpg" /> | ||
| + | </div> | ||
| + | --> | ||
| + | <div class="card border-success mb-3"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/3/30/T--SZU-China--InterLab_Flu.jpg" /> | ||
| + | </div> | ||
| + | <p class="text-center"> | ||
| + | Histogram of Different Cultures’ Fluorescence at Each Time Point | ||
| + | </p> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="indent"> | ||
| + | <h2>Colony Forming Units</h2> | ||
| + | |||
| + | <div class="card-group"> | ||
| + | <div class="card"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/1/17/T--SZU-China--InterLab_CFU_1.png" alt="Card image cap" /> | ||
| + | <div class="card-body"> | ||
| + | <div class="card-title">colonies: 1012</div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="card"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/3/37/T--SZU-China--InterLab_CFU_2.png" alt="Card image cap" /> | ||
| + | <div class="card-body"> | ||
| + | <div class="card-title">colonies: 133</div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="card"> | ||
| + | <img class="card-img-top" src="https://static.igem.org/mediawiki/2018/c/c0/T--SZU-China--InterLab_CFU_3.png" alt="Card image cap" /> | ||
| + | <div class="card-body"> | ||
| + | <div class="card-title">colonies: 17</div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="indent"> | ||
| + | <h2>Summary</h2> | ||
| + | <p>This year, according to the requirements of Measurement Committee, we measured the fluorescence data of <i>E. coli</i> strain DH5-alpha and obtained some relatively precise data. To be brief, we succeeded in reducing lab-to-lab variability in fluorescence measurements by normalizing to absolute cell count or colony-forming units (CFUs).</p> | ||
| + | </div> | ||
| + | <div class="indent"> | ||
| + | <h2>Feedback</h2> | ||
| + | <p> | ||
| + | In the protocol: <b> | ||
| + | Colony Forming Units per 0.1 OD600 E.coli cultures——Step 1: Starting Sample Preparation——2. Dilute your overnight culture to OD600...——Do This in triplicate for each culture | ||
| + | |||
| + | </b>, I think that should be added one more friendly reminder. That is, all cultures should be freezed in the ice in case the <i>E.coli</i> grows up quickly and influences the results of | ||
| + | <b>Step 3: CFU/mL/OD Calculation Instructions</b>, in which we should have obtained a good data such as 1250, 125, 12, but acutually we had got some bad data like 1150, 988, 455 at the first measurement. | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | </body> | ||
</html> | </html> | ||
Latest revision as of 04:04, 14 October 2018

InterLab Study
"Measurement is fundamentally an act of communication." - Jacob Beal
Reliable and repeatable measurement is a key component to all engineering disciplines. The same holds true for synthetic biology, which has also been called engineering biology. However, the ability to repeat measurements in different labs has been difficult. The Measurement Committee, through the InterLab study, has been developing a robust measurement procedure for green fluorescent protein (GFP) over the last several years. We chose GFP as the measurement marker for this study since it's one of the most used markers in synthetic biology and, as a result, most laboratories are equipped to measure this protein.
Calibration
OD600 Reference point
Measure the Abs600 of both the LUDOX CL-X (45% colloidal silica suspension) and water to obtain the correction factor.

Particle Standard Curve
Measure the Abs600 of a series of dilution monodisperse silica microspheres to draw the Particle Standard Curve.

Fluorescence standard curve
Measure the fluorescence of a series of diluted fluorescein to draw the fluorescein standard curve.

Cell measurement
We collaborate with the team SIAT-SCIE, because thire Distribution Kit was not delivered on time and we were during our final exam.
So we provide our kit and get the streaks of the transformed devices from them.

Histogram of Different Cultures’ Fluorescence at Each Time Point
Colony Forming Units


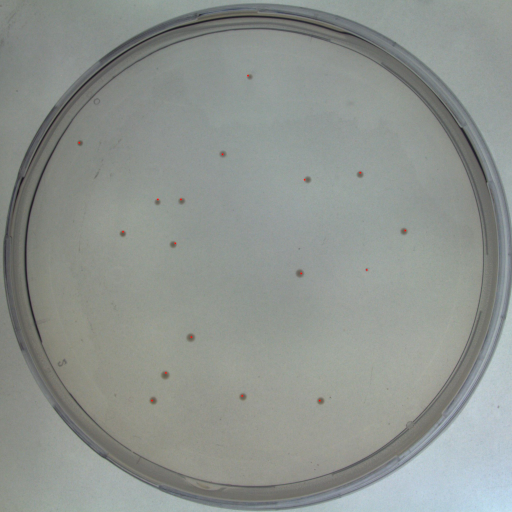
Summary
This year, according to the requirements of Measurement Committee, we measured the fluorescence data of E. coli strain DH5-alpha and obtained some relatively precise data. To be brief, we succeeded in reducing lab-to-lab variability in fluorescence measurements by normalizing to absolute cell count or colony-forming units (CFUs).
Feedback
In the protocol: Colony Forming Units per 0.1 OD600 E.coli cultures——Step 1: Starting Sample Preparation——2. Dilute your overnight culture to OD600...——Do This in triplicate for each culture , I think that should be added one more friendly reminder. That is, all cultures should be freezed in the ice in case the E.coli grows up quickly and influences the results of Step 3: CFU/mL/OD Calculation Instructions, in which we should have obtained a good data such as 1250, 125, 12, but acutually we had got some bad data like 1150, 988, 455 at the first measurement.

