| Line 129: | Line 129: | ||
</ul> | </ul> | ||
</div> | </div> | ||
| + | <div class="header"><img src="https://static.igem.org/mediawiki/2018/4/40/T--Nanjing-China--title-4.png" width="100%" onload="MM_effectAppearFade(this, 1000, 0, 100, false);MM_effectBlind('HOME', 1000, '0%', '100%', true)"></div> | ||
| + | <div class="contain"> | ||
| + | <div class="word"> | ||
| + | <div style="position:absolute; top:-90px; z-index:3; left:-10px;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/4/4a/T--Nanjing-China--PROJECT-dm.png" width="45%" /></div> | ||
| + | </div> | ||
| + | <div class="word" id="overview"> | ||
| + | <h2>Overview</h2> | ||
| + | <p>This year, we found an economic, efficient and environmental-friendly way to utilize nitrogen in the air. Our design consists of three parts: biosynthesis of CdS semiconductor, light-driven nitrogen fixation and reaction device. All three parts have gone through in-depth examination and successfully work out under real conditions.<br /> | ||
| + | The following list exhibits our key achievements in this project.<br /> | ||
| + | Constructed and tested engineered <em>E. coli</em> which express nitrogenase and OmpA-PbrR. <br /> | ||
| + | Constructed and tested light-driven system based on OmpA-PbrR protein.<br /><html xmlns="http://www.w3.org/1999/xhtml"> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | ||
| + | <title>Nanjing-China2018</title> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Nanjing-China/CSS:1?action=raw&ctype=text/css" /> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Nanjing-China/CSS:loader?action=raw&ctype=text/css" /> | ||
| + | <style type="text/css"> | ||
| + | #HQ_page table p{ font-size:85%;} | ||
| + | #HQ_page .word-note p{ text-align:center; } | ||
| + | |||
| + | </style> | ||
| + | <script type="text/javascript" src="https://2018.igem.org/Team:Nanjing-China/Javascript:1?action=raw&ctype=text/javascript"></script> | ||
| + | <script type="text/javascript"> | ||
| + | function menuFix(){ | ||
| + | var sfEles=document.getElementById("menu").getElementsByTagName("li"); | ||
| + | for(var i=0;i<sfEles.length; i++){ | ||
| + | sfEles[i].onmouseover=function(){ | ||
| + | this.className +=(this.className.length>0?"":"")+"listshow"; | ||
| + | } | ||
| + | sfEles[i].onmouseout=function(){ | ||
| + | this.className =this.className.replace("listshow",""); | ||
| + | } | ||
| + | } | ||
| + | } | ||
| + | window.onload= menuFix; | ||
| + | function MM_changeProp(objId,x,theProp,theValue) { //v9.0 | ||
| + | var obj = null; with (document){ if (getElementById) | ||
| + | obj = getElementById(objId); } | ||
| + | if (obj){ | ||
| + | if (theValue == true || theValue == false) | ||
| + | eval("obj.style."+theProp+"="+theValue); | ||
| + | else eval("obj.style."+theProp+"='"+theValue+"'"); | ||
| + | } | ||
| + | } | ||
| + | function MM_effectAppearFade(targetElement, duration, from, to, toggle) | ||
| + | { | ||
| + | Spry.Effect.DoFade(targetElement, {duration: duration, from: from, to: to, toggle: toggle}); | ||
| + | } | ||
| + | function MM_effectBlind(targetElement, duration, from, to, toggle) | ||
| + | { | ||
| + | Spry.Effect.DoBlind(targetElement, {duration: duration, from: from, to: to, toggle: toggle}); | ||
| + | } | ||
| + | </script> | ||
| + | </head> | ||
| + | |||
| + | <body> | ||
| + | <div id="loader"> | ||
| + | <div class="spinner"> | ||
| + | <div class="spinner-container container1"> | ||
| + | <div class="circle1"></div> | ||
| + | <div class="circle2"></div> | ||
| + | <div class="circle3"></div> | ||
| + | <div class="circle4"></div> | ||
| + | </div> | ||
| + | <div class="spinner-container container2"> | ||
| + | <div class="circle1"></div> | ||
| + | <div class="circle2"></div> | ||
| + | <div class="circle3"></div> | ||
| + | <div class="circle4"></div> | ||
| + | </div> | ||
| + | <div class="spinner-container container3"> | ||
| + | <div class="circle1"></div> | ||
| + | <div class="circle2"></div> | ||
| + | <div class="circle3"></div> | ||
| + | <div class="circle4"></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <script type="text/javascript"> | ||
| + | $(window).load(function() { | ||
| + | $("#loader").fadeOut("slow"); | ||
| + | }) | ||
| + | |||
| + | </script> | ||
| + | <div id="d"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/f/fe/T--Nanjing-China--signal-1.jpg" width="100%" id="Image1" onclick="MM_effectBlind('HOME', 1000, '0%', '100%', true)" /> | ||
| + | <div align="center"> | ||
| + | <div id="HOME"> | ||
| + | <div class="sub"> | ||
| + | <ul><li><a href="https://2018.igem.org/Team:Nanjing-China/Demonstrate">Demonstrate</a></li></ul></div> | ||
| + | <ul> | ||
| + | <li><a href="#overview">Overview</a></li> | ||
| + | <li><a href="#cds">CdS</a></li> | ||
| + | <li><a href="#nit"><font size="-1">Nitrogen fixation</font></a></li> | ||
| + | <li><a href="#device">Device</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div id="for_judge" align="center"><div class="i"><ul><a href="https://2018.igem.org/Team:Nanjing-China/For_Judges"><strong>For_judge</strong></a></ul></div></div> | ||
| + | <div class="container" align="center"> | ||
| + | <div id="menu" > | ||
| + | <ul> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China">N<font size="-1"><sub>2</sub></font> CHASER</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Team">Team</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Members">Members</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Attributions">Attributions</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/For_Judges">For_Judges</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Background">PROJECT</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Background">Background</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Design">Design</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Results">Results</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Demonstrate"><font size="-0.1">Demonstrate</font></a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Hardware">Hardware</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/InterLab">InterLab</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Parts">PARTS</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Basic_Part">Basic_Part</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Composite_Part">Composite</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Improve">Improve</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Model">MODELING</a></a> | ||
| + | </li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Human_Practices">PRACTICES</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Human_Practices"><font size="-1">Human_Practices</font></a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Safety">Safety</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Collaborations"><font size="-0.1">Collaboration</font></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2018.igem.org/Team:Nanjing-China/Notebook">NOTEBOOK</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
<div class="header"><img src="https://static.igem.org/mediawiki/2018/4/40/T--Nanjing-China--title-4.png" width="100%" onload="MM_effectAppearFade(this, 1000, 0, 100, false);MM_effectBlind('HOME', 1000, '0%', '100%', true)"></div> | <div class="header"><img src="https://static.igem.org/mediawiki/2018/4/40/T--Nanjing-China--title-4.png" width="100%" onload="MM_effectAppearFade(this, 1000, 0, 100, false);MM_effectBlind('HOME', 1000, '0%', '100%', true)"></div> | ||
<div class="contain"> | <div class="contain"> | ||
| Line 145: | Line 286: | ||
</div> | </div> | ||
<div class="word" id="cds"> | <div class="word" id="cds"> | ||
| − | <h3>Biosynthesis of CdS semiconductor</h3> | + | <h3>Biosynthesis of CdS semiconductor on cell surface</h3> |
<p>One key element of light-driven system in our design is CdS semiconductor precipitated by the OmpA-PbrR fused protein. We used TEM-EDX analysis to characterize CdS semiconductor precipitated on <em>E. coli</em> cell. We also determined the maximum concentration of Cd<sup>2+</sup> appropriate for strain growth, as well as the amount of CdS that can be precipitated on cell surface.</p> | <p>One key element of light-driven system in our design is CdS semiconductor precipitated by the OmpA-PbrR fused protein. We used TEM-EDX analysis to characterize CdS semiconductor precipitated on <em>E. coli</em> cell. We also determined the maximum concentration of Cd<sup>2+</sup> appropriate for strain growth, as well as the amount of CdS that can be precipitated on cell surface.</p> | ||
| − | + | <div class="word-note" align="center"><img src="https://static.igem.org/mediawiki/2018/8/8f/T--Nanjing-China--toxicity.jpg" width="70%" /> | |
| − | + | <p><font size="-1">Figure 1. Cd<sup>2+</sup> toxicity test. Cadmium ions shows no significant toxic effects on both strains.</font></p></div> | |
| − | + | ||
<div class="word-background-block" style="height:30px;"></div> | <div class="word-background-block" style="height:30px;"></div> | ||
| − | + | <div class="word-note" align="center"> | |
| + | <img src="https://static.igem.org/mediawiki/2018/0/05/T--Nanjing-China--ICP-MS.jpg" width="70%" /> | ||
| + | <p><font size="-1">Figure 2. Cd<Sup>2+</Sup> absorption test. The introduction of OmpA-PbrR confers the host cell with Cd<sup>2+</sup> absorption capacity.</font></p></div> | ||
<div class="word-background-block" style="height:30px;"></div> | <div class="word-background-block" style="height:30px;"></div> | ||
| − | + | <div class="word-note" align="center"> | |
| + | <img src="https://static.igem.org/mediawiki/2018/a/a7/T--Nanjing-China--TEX-EDX.jpg" width="100%" /> | ||
| + | <p><font size="-1">Figure 3. (a) TEM images of biosynthesized CdS nanoparticles on the surface of a EJNC cell. (b) EDX confirmation of randomly chosen CdS nanoparticle. The absorbed Cd<sup>2+</sup> precipitates on the outer membrane of EJNC in the form of CdS nanoparticles. </font></p></div> | ||
| + | <div class="word-background-block" style="height:30px;"></div> | ||
| + | <div class="word-note" align="center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/a/a9/T--Nanjing-China--UV.jpg" width="60%" /> | ||
| + | <p><font size="-1">Figure 4. Characterization of biologically precipitated CdS nanoparticles on the outer membranes of <em>E. coli</em> cells. The UV-Vis Spectrum of <em>E. coli</em>/CdS hybrids in solution demonstrating a band gap at 424 nm.</font></p> | ||
<div class="word-background-block" style="height:30px;"></div> | <div class="word-background-block" style="height:30px;"></div> | ||
| − | + | <div class="word-note" align="center"> | |
| + | <img src="https://static.igem.org/mediawiki/2018/b/be/T--Nanjing-China--Figure_S4.jpg" width="63%" /> | ||
| + | <p><font size="-1">Figure 5. Quantitative comparison of the photoelectrical capacity of in situ biosynthesized CdS nanoparticles. The concentrations of reduced methylviologen (MV) in various experimental groups confirm that the CdS nanoparticles precipitate on the EJNC cells adsorb a photon and transfer an electron to MV<sup>2+</sup>. </font></p> | ||
| + | </div> | ||
</div> | </div> | ||
<div class="word" id="nit"> | <div class="word" id="nit"> | ||
<h3>Light-driven nitrogen fixation</h3> | <h3>Light-driven nitrogen fixation</h3> | ||
<p>CdS semiconductor generate excited electrons under illumination which are then passed to nitrogenase for nitrogen fixation. We introduce nitrogenase to <em>E. coli</em> to enable it to reduce dinitrogen to ammonia. We conducted QPCR to detect relative transcriptional level of each <em>nif</em> gene. We also plan to optimize the structure of <em>nif </em>gene operon after modeling. Using acetylene reduce assay, we then verify the activity of nitrogenase in our system.</p> | <p>CdS semiconductor generate excited electrons under illumination which are then passed to nitrogenase for nitrogen fixation. We introduce nitrogenase to <em>E. coli</em> to enable it to reduce dinitrogen to ammonia. We conducted QPCR to detect relative transcriptional level of each <em>nif</em> gene. We also plan to optimize the structure of <em>nif </em>gene operon after modeling. Using acetylene reduce assay, we then verify the activity of nitrogenase in our system.</p> | ||
| − | + | <div class="word-note" align="center"> | |
| + | <img src="https://static.igem.org/mediawiki/2018/4/47/T--Nanjing-China--qRT-PCR.png" width="70%" /> | ||
| + | <p><font size="-1">Figure 6. Expression profiles of each structure gene in the <em>nif</em> cluster that overexpressed in EJNC. Relative expression compared to the housekeeping gene 16S rRNA is shown. qRT-PCR analysis demonstrates that all the component genes of the <em>nif</em> cluster are significantly over expressed in EJNC whereas the transcription of these genes are no detected (N.D.) in nondiazotrophic <em>E. coli</em> JM109. </font></p> | ||
| + | </div> | ||
</div> | </div> | ||
<div class="word" id="device"> | <div class="word" id="device"> | ||
Revision as of 06:44, 17 October 2018


Overview
This year, we found an economic, efficient and environmental-friendly way to utilize nitrogen in the air. Our design consists of three parts: biosynthesis of CdS semiconductor, light-driven nitrogen fixation and reaction device. All three parts have gone through in-depth examination and successfully work out under real conditions.
The following list exhibits our key achievements in this project.
Constructed and tested engineered E. coli which express nitrogenase and OmpA-PbrR.
Constructed and tested light-driven system based on OmpA-PbrR protein.


Overview
This year, we found an economic, efficient and environmental-friendly way to utilize nitrogen in the air. Our design consists of three parts: biosynthesis of CdS semiconductor, light-driven nitrogen fixation and reaction device. All three parts have gone through in-depth examination and successfully work out under real conditions.
The following list exhibits our key achievements in this project.
Constructed and tested engineered E. coli which express nitrogenase and OmpA-PbrR.
Constructed and tested light-driven system based on OmpA-PbrR protein.
Achieved light-driven nitrogen fixation.
Designed a reaction device that cater for our light-driven nitrogen fixation reaction.
Biosynthesis of CdS semiconductor on cell surface
One key element of light-driven system in our design is CdS semiconductor precipitated by the OmpA-PbrR fused protein. We used TEM-EDX analysis to characterize CdS semiconductor precipitated on E. coli cell. We also determined the maximum concentration of Cd2+ appropriate for strain growth, as well as the amount of CdS that can be precipitated on cell surface.

Figure 1. Cd2+ toxicity test. Cadmium ions shows no significant toxic effects on both strains.

Figure 2. Cd2+ absorption test. The introduction of OmpA-PbrR confers the host cell with Cd2+ absorption capacity.
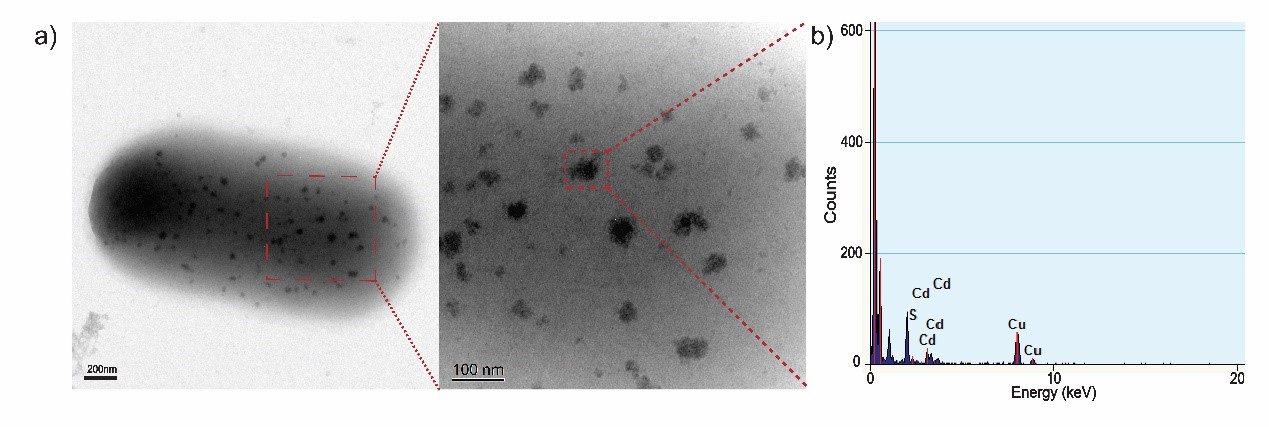
Figure 3. (a) TEM images of biosynthesized CdS nanoparticles on the surface of a EJNC cell. (b) EDX confirmation of randomly chosen CdS nanoparticle. The absorbed Cd2+ precipitates on the outer membrane of EJNC in the form of CdS nanoparticles.

Figure 4. Characterization of biologically precipitated CdS nanoparticles on the outer membranes of E. coli cells. The UV-Vis Spectrum of E. coli/CdS hybrids in solution demonstrating a band gap at 424 nm.

Figure 5. Quantitative comparison of the photoelectrical capacity of in situ biosynthesized CdS nanoparticles. The concentrations of reduced methylviologen (MV) in various experimental groups confirm that the CdS nanoparticles precipitate on the EJNC cells adsorb a photon and transfer an electron to MV2+.
Light-driven nitrogen fixation
CdS semiconductor generate excited electrons under illumination which are then passed to nitrogenase for nitrogen fixation. We introduce nitrogenase to E. coli to enable it to reduce dinitrogen to ammonia. We conducted QPCR to detect relative transcriptional level of each nif gene. We also plan to optimize the structure of nif gene operon after modeling. Using acetylene reduce assay, we then verify the activity of nitrogenase in our system.

Figure 6. Expression profiles of each structure gene in the nif cluster that overexpressed in EJNC. Relative expression compared to the housekeeping gene 16S rRNA is shown. qRT-PCR analysis demonstrates that all the component genes of the nif cluster are significantly over expressed in EJNC whereas the transcription of these genes are no detected (N.D.) in nondiazotrophic E. coli JM109.
Reaction device
We designed a light-driven microbe reaction device to apply our system to practical use. After a few test, we proved our device to be quite practical. (see device for more details)







