(Prototype team page) |
Zhaowenxue (Talk | contribs) |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
<html> | <html> | ||
| + | |||
| + | <div class="container" id="section-2"> | ||
| + | <div class="paragraph shadow"> | ||
| + | <h2 class="title">experiment Results</h2> | ||
| + | <h3 class="title">promoter library construction</h3> | ||
| + | <p>Through our wet lab work, we have got 23 promoter strength data. The measured values are as follows:</p> | ||
| − | < | + | <span class="image fit"> |
| − | < | + | <img src="https://static.igem.org/mediawiki/2018/c/c9/T--SKLMT-China--projectresultpromoter.png |
| − | <p> | + | " alt="Table.1 promoter strength characterized by a reporter gene" /> |
| − | </ | + | </span> |
| + | <p>The fluorescence from the luciferase gene without a promoter and from <latin>P.fluorescence pf-5</latin>, that contains no luciferase gene, was also determined. Since predecessors have less research on the promoter collection and characterization of<latin> P. fluorescence</latin>, we have done an innovative work and set a standard for the promoter strength in <latin>P.fluorescence</latin>.We set <latin>P16s </latin>promoter as the standard, so the activity of the promoters was compared to the activity of the<latin> 13P16s</latin> promoter in our project. Some of the promoters which resulted from this approach, turned out to be very strong (more than 7-fold the <latin>P16s</latin> promoter), others quite weak (almost 0.5-fold lower than the <latin>P16s </latin>promoter)<p> | ||
| + | <p>Nowadays, the focus in metabolic engineering research is shifting from massive overexpression and inactivation of genes towards the model-based fine tuning of gene expression. In other words, being able to rationally designing a promoter would be extremely profitable in the context of a model-based metabolic engineering. Our project therefore attempts to link the promoter sequence to its strength. To this end, modelling strategies have been applied. (To learn more, please read our Model )<p> | ||
| + | <p>By solving the matrix sparse solution algorithm, we can conclude that there is indeed a linear relationship between the total 64 data of AAA-GGG and the promoter strength. Notice that we used merely 22 sets of data to approximately solve the sparse solutions of 64 equations with a fitting precision higher than 95%, therefore, when we provide as much data as possible, the fitting precision of the model will greatly increase.<p> | ||
| + | <p>We notice that there is a significant difference in the magnitude of the measurements between our group and the other groups. In order to better contribute our results to other teams, we will use the magnitude of our measurements and classify the promoters by numerical size.<p> | ||
| + | <p>The promoter intensity level corresponding to the data is as follows:</p> | ||
| + | <span class="image fit"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/8/89/T--SKLMT-China--promterleveleqution.png" alt=" " /> | ||
| + | </span> | ||
| − | < | + | <p>Using this standard, we quickly categorized 23 known promoters:</p> |
| − | < | + | <span class="image fit"> |
| − | + | <img src="https://static.igem.org/mediawiki/2018/6/6c/T--SKLMT-China--promoterstrengthfig.png | |
| − | + | " alt=" Fig.1 The strength level of different promoters" /> | |
| − | + | </span> | |
| − | + | ||
| − | + | ||
| − | </ | + | |
| + | </div> | ||
| + | </div> | ||
| + | <div class="container" id="section-2"> | ||
| − | <div class=" | + | <div class="paragraph shadow"> |
| − | < | + | <h2 class="title">Demonstrate</h2> |
| − | < | + | <p>Based on our promoter library, we selected three promoters (promoter 4, promoter 5 and promoter11) of different intensities to regulate the expression of the key nicotine-degrading gene <i>nicA2</i>.We’ve constructed lasmid, pBBR1-km-amp-cm-promoter-nic, and electroporated it into our new chassis bacteria,<latin> Pseudomonas fluorescences pf-5</latin>. </p> |
| − | + | <span class="image fit"> | |
| − | < | + | <img src="https://static.igem.org/mediawiki/2018/2/20/T--SKLMT-China--demonstrationfig1.png |
| − | < | + | "alt=" "width=450 height=500/> |
| − | + | </span> | |
| − | </ | + | |
| − | < | + | <p>We found that there are only a few colonies of <latin> P. fluorescences pf-5</latin> (containing <i>nicA2 </i>with promoters in front of it)on the LB plate,but lots of colonies on the <i>CK </i>plate(control group electroporated with pBBR1-km-amp-cm-nic without promoter). Combining the fact that theE.coliusually lost a part of (sometimes the whole plasmid)when we want to use high-copy vector to carry this gene cluster,we wondered if it<i> NicA2 </i>is a poisonous enzyme so that <latin>E.coli </latin>lost this plasmid during DNA replication under stress. </p> |
| + | <div style="top: 0px;"></div><span style="font-family:宋体;font-size:16px;"></span><table style="background: white; border: currentColor; border-image: none; border-collapse: collapse; mso-yfti-tbllook: 1184; mso-padding-alt: 0cm 5.4pt 0cm 5.4pt; mso-border-top-alt: solid black 1.0pt; mso-background-themecolor: background1; mso-border-top-themecolor: text1; mso-border-bottom-alt: solid black 1.0pt; mso-border-bottom-themecolor: text1;" border="1" cellspacing="0" cellpadding="0"><span style="font-family:宋体;"> </span><tbody><tr style="mso-yfti-irow: -1; mso-yfti-firstrow: yes;"><span style="font-family:宋体;"> </span><td width="304" valign="top" style="border-width: 1pt 0px; border-style: solid none; border-color: black rgb(0, 0, 0); padding: 0cm 5.4pt; width: 182.6pt; background-color: transparent; mso-border-top-themecolor: text1; mso-border-bottom-themecolor: text1;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 5;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">Plasmid in </span></span><em><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; font-size: 10pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;">P. fluorescences </span></em><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; font-size: 10pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold; mso-bidi-font-style: italic;">pf-5</span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="213" valign="top" style="border-width: 1pt 0px; border-style: solid none; border-color: black rgb(0, 0, 0); padding: 0cm 5.4pt; width: 127.6pt; background-color: transparent; mso-border-top-themecolor: text1; mso-border-bottom-themecolor: text1;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 1;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">Strength of promoter</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="193" valign="top" style="border-width: 1pt 0px; border-style: solid none; border-color: black rgb(0, 0, 0); padding: 0cm 5.4pt; width: 115.9pt; background-color: transparent; mso-border-top-themecolor: text1; mso-border-bottom-themecolor: text1;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 1;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">CFU on LB plate</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span></tr><span style="font-family:宋体;"> </span><tr style="mso-yfti-irow: 0;"><span style="font-family:宋体;"> </span><td width="304" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 182.6pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 68;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">pBBR1-km-amp-cm-nic</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="213" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 127.6pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 64;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">none</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="193" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 115.9pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 64;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">>200</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span></tr><span style="font-family:宋体;"> </span><tr style="mso-yfti-irow: 1;"><span style="font-family:宋体;"> </span><td width="304" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 182.6pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 4;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">pBBR1-km-amp-cm-promoter4-nic</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="213" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 127.6pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">normal</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="193" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 115.9pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">5</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span></tr><span style="font-family:宋体;"> </span><tr style="mso-yfti-irow: 2;"><span style="font-family:宋体;"> </span><td width="304" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 182.6pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 68;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">pBBR1-km-amp-cm-promoter5-nic</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="213" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 127.6pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 64;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">weak</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="193" valign="top" style="padding: 0cm 5.4pt; border: 0px rgb(0, 0, 0); border-image: none; width: 115.9pt; background-color: transparent;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 64;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">5</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span></tr><span style="font-family:宋体;"> </span><tr style="mso-yfti-irow: 3; mso-yfti-lastrow: yes;"><span style="font-family:宋体;"> </span><td width="304" valign="top" style="border-width: 0px 0px 1pt; border-style: none none solid; border-color: rgb(0, 0, 0) rgb(0, 0, 0) black; padding: 0cm 5.4pt; border-image: none; width: 182.6pt; background-color: transparent; mso-border-bottom-themecolor: text1;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center; mso-yfti-cnfc: 4;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191; mso-bidi-font-weight: bold;"><span style="font-size:16px;">pBBR1-km-amp-cm-promoter11-nic</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="213" valign="top" style="border-width: 0px 0px 1pt; border-style: none none solid; border-color: rgb(0, 0, 0) rgb(0, 0, 0) black; padding: 0cm 5.4pt; border-image: none; width: 127.6pt; background-color: transparent; mso-border-bottom-themecolor: text1;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">strong</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span><td width="193" valign="top" style="border-width: 0px 0px 1pt; border-style: none none solid; border-color: rgb(0, 0, 0) rgb(0, 0, 0) black; padding: 0cm 5.4pt; border-image: none; width: 115.9pt; background-color: transparent; mso-border-bottom-themecolor: text1;"><span style="font-family:宋体;font-size:16px;"> </span><p align="center" style="margin: 0cm 0cm 0pt; text-align: center;"><span lang="EN-US" style="color: black; font-family: 'Times New Roman','serif'; mso-bidi-font-size: 10.5pt; mso-fareast-font-family: 仿宋; mso-themecolor: text1; mso-themeshade: 191;"><span style="font-size:16px;">1</span></span></p><span style="font-family:宋体;font-size:16px;"> </span></td><span style="font-family:宋体;"> </span></tr><span style="font-family:宋体;"></span></tbody></table><span style="font-family:宋体;font-size:16px;"></span> | ||
| − | < | + | <p>In order to test if our plasmids really work in <latin> P. fluorescences pf-5</latin>, we demonstrate our project in three levels, transcription, protein expression, and substrate degradation efficiency. </p> |
| − | < | + | <p>We used <i>SDS-PAGE</i>, to demonstrate the expression of <i>NicA2</i> (52.5 kDa). In the picture,we can see the clear expression of <i>NicA2 </i>in <latin> P. fluorescences pf-5</latin> containing plasmid with promoter. It can be qualitatively known that our promoter can initiate transcription normally, and the nicotine-degrading gene cluster has also been successfully heterologously expressed.</p> |
| − | < | + | <span class="image fit"> |
| − | + | <img src="https://static.igem.org/mediawiki/2018/e/ea/T--SKLMT-China--demonstrationfig2.png" alt="Fig.2 SDS-PAGE result of protein NicA2 " /> | |
| − | + | </span> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
| + | <p>As for transcription, we are going to use real-time <i>PCR</i> to compare the ability ofinitiating transcription of three promoters .</p> | ||
| + | <p>In addition, we are comparing the degradation efficiency by<i> HPLC</i>. <i>NicA2</i> in can convert nicotine to pseudo-oxidation in a whole-cell reaction, and the rest of the gene cluster will convert pseudo-oxidation to 2,5-<i>DHP</i>. </p> | ||
| + | <p>However, due to time limitation, we were unable to complete the last two experiments, but we will do a supplementary explanation in our presentation. Please stay tuned. </p> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
</html> | </html> | ||
| + | {{SKLMT-China/footer}} | ||
Latest revision as of 11:27, 17 October 2018
experiment Results
promoter library construction
Through our wet lab work, we have got 23 promoter strength data. The measured values are as follows:

The fluorescence from the luciferase gene without a promoter and from
Nowadays, the focus in metabolic engineering research is shifting from massive overexpression and inactivation of genes towards the model-based fine tuning of gene expression. In other words, being able to rationally designing a promoter would be extremely profitable in the context of a model-based metabolic engineering. Our project therefore attempts to link the promoter sequence to its strength. To this end, modelling strategies have been applied. (To learn more, please read our Model )
By solving the matrix sparse solution algorithm, we can conclude that there is indeed a linear relationship between the total 64 data of AAA-GGG and the promoter strength. Notice that we used merely 22 sets of data to approximately solve the sparse solutions of 64 equations with a fitting precision higher than 95%, therefore, when we provide as much data as possible, the fitting precision of the model will greatly increase.
We notice that there is a significant difference in the magnitude of the measurements between our group and the other groups. In order to better contribute our results to other teams, we will use the magnitude of our measurements and classify the promoters by numerical size.
The promoter intensity level corresponding to the data is as follows:

Using this standard, we quickly categorized 23 known promoters:

Demonstrate
Based on our promoter library, we selected three promoters (promoter 4, promoter 5 and promoter11) of different intensities to regulate the expression of the key nicotine-degrading gene nicA2.We’ve constructed lasmid, pBBR1-km-amp-cm-promoter-nic, and electroporated it into our new chassis bacteria,
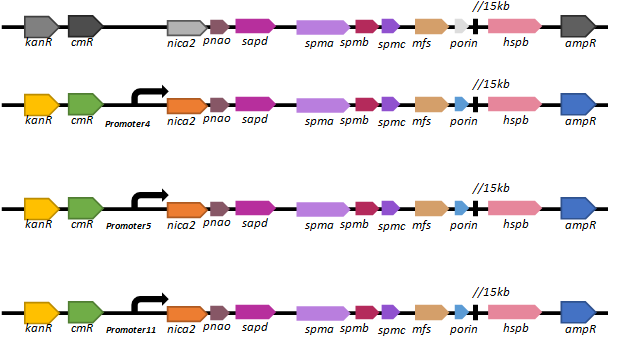
We found that there are only a few colonies of
| Plasmid in P. fluorescences pf-5 | Strength of promoter | CFU on LB plate |
| pBBR1-km-amp-cm-nic | none | >200 |
| pBBR1-km-amp-cm-promoter4-nic | normal | 5 |
| pBBR1-km-amp-cm-promoter5-nic | weak | 5 |
| pBBR1-km-amp-cm-promoter11-nic | strong | 1 |
In order to test if our plasmids really work in
We used SDS-PAGE, to demonstrate the expression of NicA2 (52.5 kDa). In the picture,we can see the clear expression of NicA2 in

As for transcription, we are going to use real-time PCR to compare the ability ofinitiating transcription of three promoters .
In addition, we are comparing the degradation efficiency by HPLC. NicA2 in can convert nicotine to pseudo-oxidation in a whole-cell reaction, and the rest of the gene cluster will convert pseudo-oxidation to 2,5-DHP.
However, due to time limitation, we were unable to complete the last two experiments, but we will do a supplementary explanation in our presentation. Please stay tuned.
