Zhaowenxue (Talk | contribs) |
Zhaowenxue (Talk | contribs) |
||
| Line 8: | Line 8: | ||
<span class="image fit"> | <span class="image fit"> | ||
<img src="https://static.igem.org/mediawiki/2018/2/20/T--SKLMT-China--demonstrationfig1.png | <img src="https://static.igem.org/mediawiki/2018/2/20/T--SKLMT-China--demonstrationfig1.png | ||
| − | " | + | "width=700 height=650/> |
</span> | </span> | ||
| Line 19: | Line 19: | ||
<p>In order to test if our plasmids really work in <latin> P. fluorescences pf-5</latin>, we demonstrate our project in three levels, transcription, protein expression, and substrate degradation efficiency. </p> | <p>In order to test if our plasmids really work in <latin> P. fluorescences pf-5</latin>, we demonstrate our project in three levels, transcription, protein expression, and substrate degradation efficiency. </p> | ||
<p>We used <i>SDS-PAGE</i>, to demonstrate the expression of <i>NicA2</i> (52.5 kDa). In the picture,we can see the clear expression of <i>NicA2 </i>in <latin> P. fluorescences pf-5</latin> containing plasmid with promoter. It can be qualitatively known that our promoter can initiate transcription normally, and the nicotine-degrading gene cluster has also been successfully heterologously expressed</p> | <p>We used <i>SDS-PAGE</i>, to demonstrate the expression of <i>NicA2</i> (52.5 kDa). In the picture,we can see the clear expression of <i>NicA2 </i>in <latin> P. fluorescences pf-5</latin> containing plasmid with promoter. It can be qualitatively known that our promoter can initiate transcription normally, and the nicotine-degrading gene cluster has also been successfully heterologously expressed</p> | ||
| − | <p>As for transcription, we are going to use real-time <i>PCR</i> to compare the ability ofinitiating transcription of three promoters. </p> | + | <p>As for transcription, we are going to use real-time <i>PCR</i> to compare the ability ofinitiating transcription of three promoters. </p>In addition, we are comparing the degradation efficiency by<i> HPLC</i>. <i>NicA2</i> in can convert nicotine to pseudo-oxidation in a whole-cell reaction, and the rest of the gene cluster will convert pseudo-oxidation to 2,5-<i>DHP</i>. </p> |
| − | In addition, we are comparing the degradation efficiency by<i> HPLC</i>. <i>NicA2</i> in can convert nicotine to pseudo-oxidation in a whole-cell reaction, and the rest of the gene cluster will convert pseudo-oxidation to 2,5-<i>DHP</i>. </p> | + | |
<p>However, due to time limitation, we were unable to complete the last two experiments, but we will do a supplementary explanation in our presentation. Please stay tuned. </p> | <p>However, due to time limitation, we were unable to complete the last two experiments, but we will do a supplementary explanation in our presentation. Please stay tuned. </p> | ||
Revision as of 10:19, 17 October 2018
Demonstrate
Based on our promoter library, we selected three promoters (promoter 4, promoter 5 and promoter11) of different intensities to regulate the expression of the key nicotine-degrading gene nicA2.We’ve constructed lasmid, pBBR1-km-amp-cm-promoter-nic, and electroporated it into our new chassis bacteria,
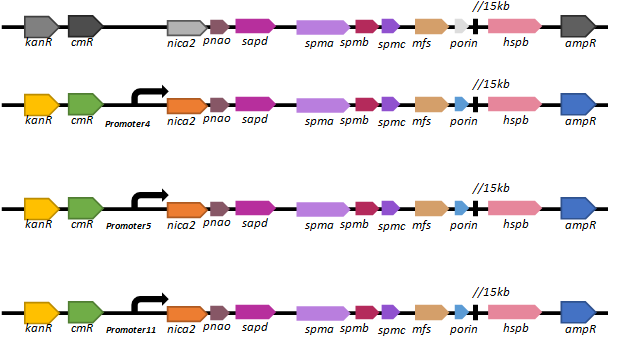
We found that there are only a few colonies of
| Plasmid in P. fluorescences pf-5 | Strength of promoter | CFU on LB plate |
| pBBR1-km-amp-cm-nic | none | >200 |
| pBBR1-km-amp-cm-promoter4-nic | normal | 5 |
| pBBR1-km-amp-cm-promoter5-nic | weak | 5 |
| pBBR1-km-amp-cm-promoter11-nic | strong | 1 |
In order to test if our plasmids really work in
We used SDS-PAGE, to demonstrate the expression of NicA2 (52.5 kDa). In the picture,we can see the clear expression of NicA2 in
As for transcription, we are going to use real-time PCR to compare the ability ofinitiating transcription of three promoters.
In addition, we are comparing the degradation efficiency by HPLC. NicA2 in can convert nicotine to pseudo-oxidation in a whole-cell reaction, and the rest of the gene cluster will convert pseudo-oxidation to 2,5-DHP.However, due to time limitation, we were unable to complete the last two experiments, but we will do a supplementary explanation in our presentation. Please stay tuned.
