
Cell system

How can we design a screening platform for potential AGA drugs?
AGA has been shown to highly correlate with the amount of DKK1 protein produced in hair follicles.[1] Therefore, we selected this specific pathway for drug screening. Our platform aims to monitor the level of DKK1 protein secretion in vitro via both Dermal Papilla and HEK 293T cell lines. Molecules that are able to inhibit the DKK1 protein secretion pathway in hair follicles may become candidate drugs for AGA therapy.
How do we select our model cell lines?
The goal of our project is to establish a high throughput drug screening platform that can approximately model the DKK1 molecular pathway of hair follicles. Therefore, DP cell line is chosen as our primary model cells as DP cells play an important role in hair follicle morphogenesis and regeneration [2](Figure 1). However, DP cells are relatively expensive and have considerably longer life cycle compared to commonly used cell lines such as HEK 293 and Hep G2[3]. This contradicts with our original idea of developing a cheap and easy model for drug screening. Therefore, we decided to use another cell line, HEK 293T, as our second model. HEK 293T, an immortalized kidney cancer cell line, also has androgen-responsive pathway as in DP cells, but is much cheaper and easier to handle and is therefore a proper cell model for our project.

Taipei Veterans General Hospital Department of Dermatology generously sponsored us with materials related with our mammalian cell experiments, allowing us to experiment with both of the cell lines mentioned above in our platform. We deeply appreciate their help.
How can our cells detect the concentration of DKK1?
To detect the amount of DKK1 protein secreted by cells, our team decided to design a plasmid that enables mammalian cells to produce fluorescent protein in quantities proportionate to the amount of DKK1 synthesized by the cell. Both DP and HEK293T cell lines possess androgen-responsive pathway and have functional DKK1 promoter as well as DKK1 structure gene. Therefore, we constructed a plasmid with an insert consisting a mCherry protein gene under the control of a DKK1-promoter (BBa_K27510015) as shown in Figure 2. The plasmid is then transfected into both DP and HEK293T cells. We expect the mCherry to function as a reporter upon the activation of DKK1 promoter.
How do we standardize the amount of plasmid transfected into the cells?
During the process of transfection, variables such as transfection efficiency, cell number, and well position can interfere with experimental results. Therefore we designed an internal control plasmid (BBa_K2751013) consisting an mEGFP fluorescence protein (BBa_K2751011) under the control of a CMV promoter. The CMV promoter allows mEGFP to be secreted continuously when transfected into a cell. From detecting the internal control, we could take different transfection conditions into account and standardize all data collected[4].

How can we detect the fluorescence protein?
The most standard way of visualizing fluorescence proteins is to use a fluorescence microscope. Considering that the fluorescence proteins are expressed in cells, we choose to use an inverted confocal microscope to observe the secreted fluorescence proteins. However, this method takes a long time and requires a high quality machine. Therefore, we added an albumin secreting peptide (ALB) to mCherry. This way, mCherry will be secreted into the extracellular domain, which is more convenient for detection purposes. We could then use a plate reader to determine the amount of secreted fluorescence proteins.
References
- Inui, S., & Itami, S. Molecular basis of androgenetic alopecia: From androgen to paracrine mediators through dermal papilla. J Dermatol Sci. 2011 Jan;61(1):1-6.
- Veraitch, O., et al., Induction of hair follicle dermal papilla cell properties in human induced pluripotent stem cell-derived multipotent LNGFR(+)THY-1(+) mesenchymal cells. Sci Rep. 2017 Feb 21;7:42777.
- Cesare Achilli, Annarita Ciana, Giampaolo Minetti.Immortalized HEK 293T kidney cell lines as models of renal cells: friends or foes? J Controversies Biomed Res. 2018;4(1):6–9.
- Rodriguez-Mulero, S., & Montanya, E. Selection of a Suitable Internal Control Gene for Expression Studies in Pancreatic Islet Grafts. Transplantation,2005, 80(5), 650–652.
FRET system
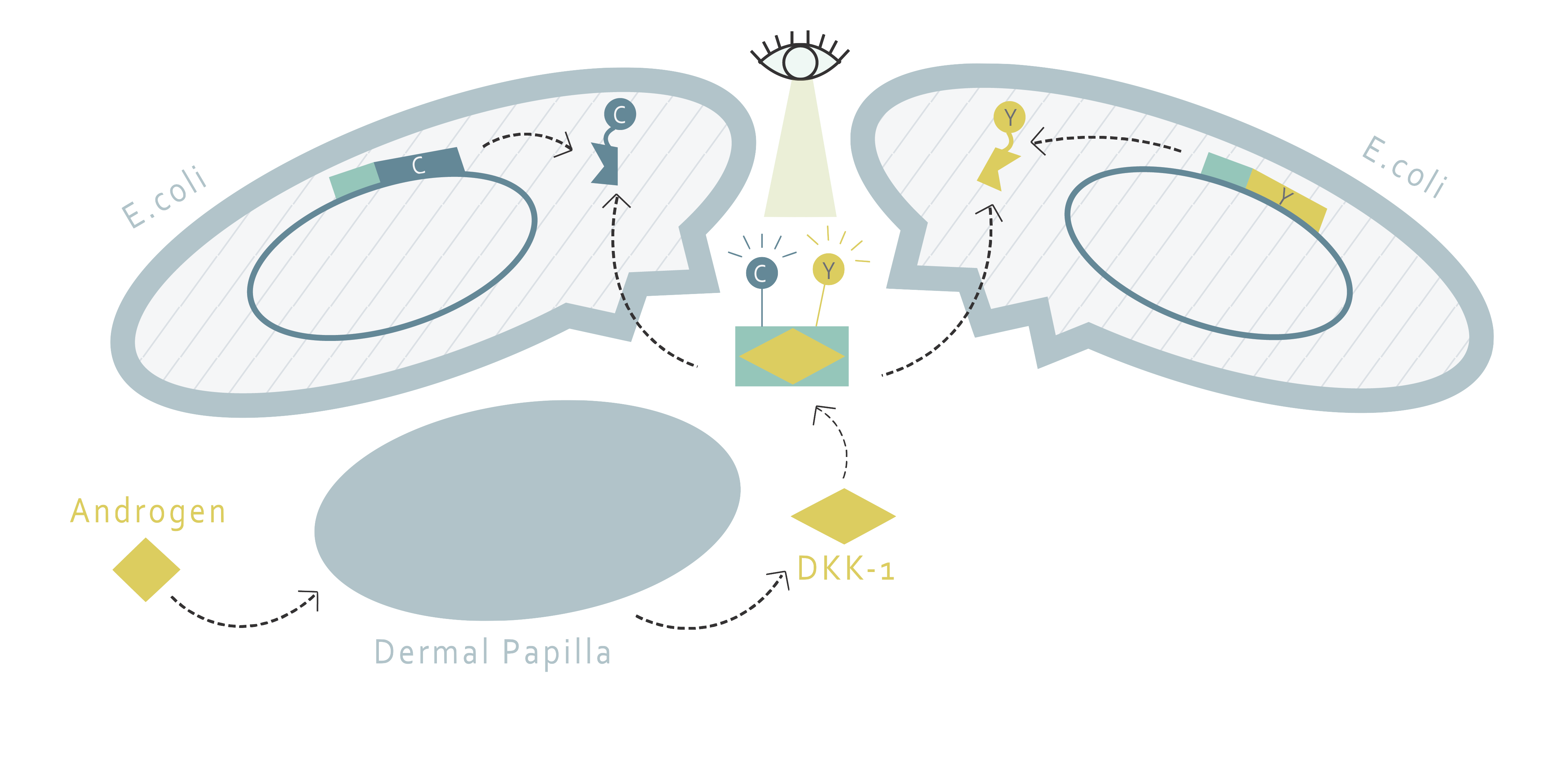
How do we establish a drug screening system without creating a new cell line ?
This part of our project aims to determine the amount of DKK1 in cell supernatants through fluorescence resonance energy transfer (FRET). FRET requires two molecules: a donor and an acceptor. If the emission wavelength of the donor molecule and the absorption wavelength of the acceptor molecule overlap each other, the excitation of the donor molecule will result in the fluorescence emission of the acceptor molecule when the two molecules are nearby. Therefore, the FRET system allows us to determine whether or not two molecules are close, or most likely, interacting with each other.

Using this method, we would only need to find a FRET pair and two proteins that bind to DKK1 and fusion them in the form of one FRET protein to one binding protein. This fusion pair can be produced by E.coli and purified through protein purification. Afterwards, with proper buffer, one would only have to add the fusion proteins into the supernatant of cells and observe the fluorescence pattern to determine the amount of DKK1.
How can we bind the FRET pairs to DKK1?
Our design requires two proteins to interact with DKK1 simultaneously by binding to different binding sites on DKK1. There are several potential candidate proteins that we can choose from. G5 and H7 are nanobodies that have been proven to bind to DKK1[1]. The other option is the low-density lipoprotein receptor-related protein 6 (LRP6). LRP6 is a protein involved in the Wnt pathway and is regulated by DKK1 binding. It contains four subdomains, including four EGF-like domains (E1-4). However, the size of the whole LRP6 protein is too big and is not suitable for FRET purposes. According to literature, dividing LRP6 into LRP6-E1E2 and LRP6-E3E4 does not interfere with their abilities to bind to DKK1[2]. Moreover, LRP6-E1 and LRP6-E3 propellers are suspected to play a crucial role in DKK1 binding. The best protein pair for DKK1 binding is determined by experiment.

What FRET pairs should we use?

For the FRET pair, we have chosen CyPet and YPet. The FRET pair is a cyan fluorescent protein and yellow fluorescent protein optimized for FRET reactions. After deciding the protein pair to use, each of the protein will be fused with CyPet or YPet, a fluorescent protein pair developed with enhanced FRET efficiency [3]. The peak excitation and emission wavelengths of CyPet and YPet are 414 and 475nm, 515 and 530nm, respectively [4]. Something to notice is that in order to minimize the destruction on protein structure during purification, the His tag is added to the fluorescent protein instead of the binding protein. This leads us to the design of two kinds of plasmids shown below:
How do we determine the amount?
When DKK1 is present, we hypothesize that the selected proteins will both bind to DKK1, bringing CyPet and YPet close enough for FRET to happen. By detecting the strength of fluorescence emission, we can thereby determine the amount of DKK1 proteins existing in the supernatant.
References
- Zhong, L., et al. Endogenous DKK1 and FRZB Regulate Chondrogenesis and Hypertrophy in Three-Dimensional Cultures of Human Chondrocytes and Human Mesenchymal Stem Cells.Stem Cells Dev. 2016 Dec 1; 25(23): 1808–1817.
- Cheng, Z., et al., Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol. 2011 Oct 9;18(11):1204-10.
- Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005 Mar;23(3):355-60.
- https://www.fpbase.org/protein/cypet/, https://www.fpbase.org/protein/ypet/

