| Line 7,338: | Line 7,338: | ||
<br> | <br> | ||
</div> | </div> | ||
| − | + | <br> | |
<div class="row" style="padding-left:50px; padding-right: 50px;"> | <div class="row" style="padding-left:50px; padding-right: 50px;"> | ||
Revision as of 04:27, 16 October 2018

Part Collection:
Transcription-activator-like effector (TALE) Stabilised Promoter Family
Transcription-activator-like-effector stabilised promoters (TALEsp) are a type of promoters able to untie gene expression level from gene copy number. They differ in the TALE used for repression and the core promoter which the TALE represses. In this study, we have used two TALE proteins reported previously: TALE1 and TALE2. Through mutating core promoters of existing pTALEsp, and adding TALE binding sites to the classical consensus promoter J23119, we created six more pTALEsp, and have added all of twelve pTALEsps to the iGEM Registry.


TALE1:
- TALE1 sp1 (http://parts.igem.org/Parts:BBa_K2753030)
- TALE1 sp2 (http://parts.igem.org/Parts:BBa_K2753024)
- TALE1 sp3 (http://parts.igem.org/Parts:BBa_K2753025)
- TALE1 sp4 (http://parts.igem.org/Parts:BBa_K2753026)
- TALE1 sp5 (http://parts.igem.org/Parts:BBa_K2753027)
- TALE1 sp6 (http://parts.igem.org/Parts:BBa_K2753029)
TALE2:
- TALE2 sp1 (http://parts.igem.org/Parts:BBa_K2753018)
- TALE2 sp2 (http://parts.igem.org/Parts:BBa_K2753019)
- TALE2 sp3 (http://parts.igem.org/Parts:BBa_K2753020)
- TALE2 sp4 (http://parts.igem.org/Parts:BBa_K2753021)
- TALE2 sp5 (http://parts.igem.org/Parts:BBa_K2753022)
- TALE2 sp6 (http://parts.igem.org/Parts:BBa_K2753023)
Table. 1 The source of all promoters in the library

All promoters are placed on the standard biobrick assembly compatible backbone pSB1C3, with a RBS (BBa_B0034) reporter gene sfGFP (BBa_K1679038) downstream of the promoters in between the restriction sites of SpeI and PstI. So teams can change reporter genes as they wish.

For TALE stabilised promoters repressed by the same TALE, the only difference between them is the sequence of the core promoter which the TALE binds to. By replacing this promoter, new TALE stabilised promoters could then be easily created. For making the process more convenient, we designed this composite part in which a genetic circuit encoding for amilCP, a blue pigment, replaces the core promoter sequence, and downstream are a RBS, a coding sequence of sfGFP, and a terminator. Two restriction sites of BsaI are put on the two ends of the amilCP circuit. By adding the same digestion sites on the promoter to be assembled, the amilCP circuit could be substituted by Golden Gate. The blue colour of amilCP and green colour of sfGFP provide a rapid distinguishment of the successful construct. If the amilCP circuit is not replaced, the colony grown would be blue as amilCP is being expressed. But if a promoter sequence substitutes amilCP circuit, sfGFP would be expressed instead of amilCP, making the colony fluorescent and easy to recognise.
Except for TALE1sp1 and TALE2sp1 which were synthesized de novo by Genscript, we assembled the rest of TALEsps by first obtaining core promoters using SOE PCR then replacing the amilCP circuit with them.
Characterization
In order to verify the stabilization effect of the pTALE promoter family, we assembled all promoter members onto three backbones:
pUC20: copy number ≈ 500
pR6K: copy number ≈ 15
pSC101: copy number ≈ 1
An identical sfGFP is placed downstream of all promoters as a reporter gene. Three constitutive promoters: J23119, J23101, J23105, with the same sfGFP downstream were characterized along with the pTALE promoters as a reference of promoter strength. The green fluoresce was measured by flow cytometry. (see methods in our notebook).

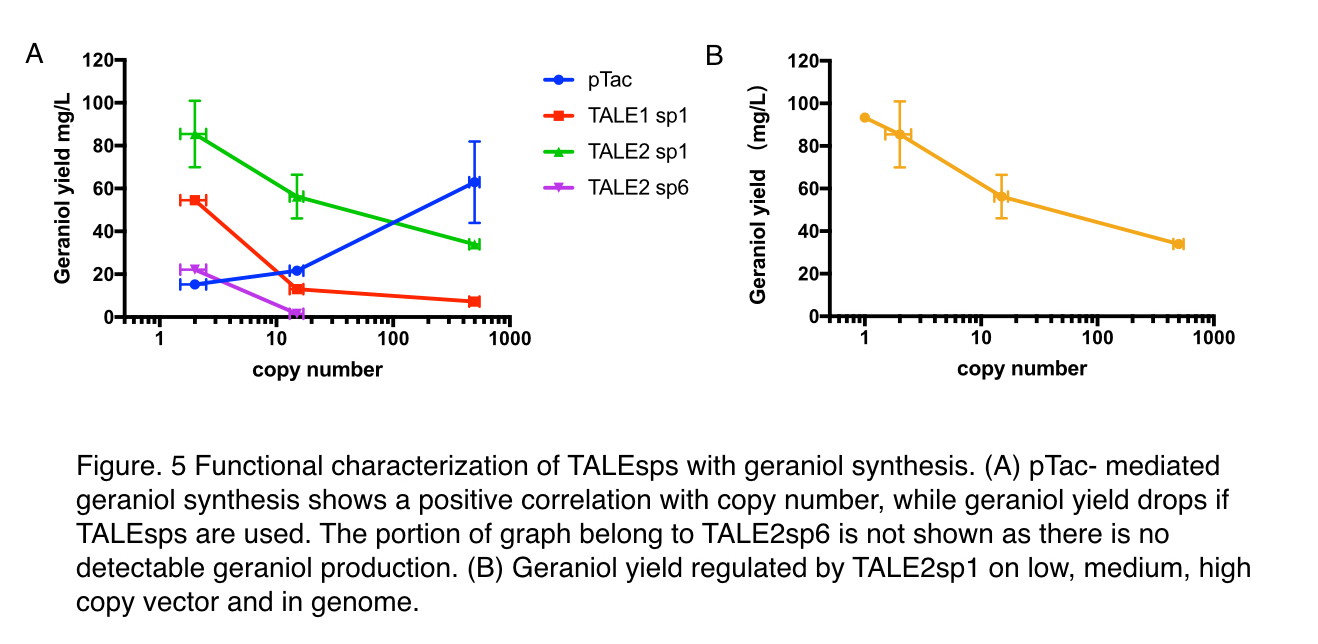
To decide whether pTALE can perform as expected in metabolic pathways, maintaining the pre-set ratio of production enzymes for ensuring optimal flux to the product, we tested pTALE1 sp1 and pTALE2 sp1 by comparison to pTac, a frequently-used IPTG inducible promoter, using an geraniol synthesis operon (Part:BBa_K2753015) containing a GPPS (Part:BBa_K2753002) and a GES (Part:BBa_K2753003). Similar to the characterisation of green fluoresce characterization, three backbones: pUC20, pR6K, pSC101 are used.