Cyclohexane (Talk | contribs) |
Cyclohexane (Talk | contribs) |
||
| (63 intermediate revisions by 6 users not shown) | |||
| Line 23: | Line 23: | ||
<meta name="msapplication-tap-highlight" content="no"><!-- windows phone 点击无高光 --> | <meta name="msapplication-tap-highlight" content="no"><!-- windows phone 点击无高光 --> | ||
<title>Team:XMU-China/Design - 2018.igem.org</title> | <title>Team:XMU-China/Design - 2018.igem.org</title> | ||
| − | <link rel="stylesheet" href="css/ | + | <link rel="stylesheet" href="css/description.css"> |
<link href="http://cdn.bootcss.com/font-awesome/4.7.0/css/font-awesome.min.css" rel="stylesheet"> | <link href="http://cdn.bootcss.com/font-awesome/4.7.0/css/font-awesome.min.css" rel="stylesheet"> | ||
<link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/cover?action=raw&ctype=text/css"> | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/cover?action=raw&ctype=text/css"> | ||
| Line 29: | Line 29: | ||
<link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/nav?action=raw&ctype=text/css"> | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/nav?action=raw&ctype=text/css"> | ||
<link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/nav_mobile?action=raw&ctype=text/css"> | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/nav_mobile?action=raw&ctype=text/css"> | ||
| − | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/ | + | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/description?action=raw&ctype=text/css"> |
<link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/font?action=raw&ctype=text/css"> | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/font?action=raw&ctype=text/css"> | ||
<link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/material-scrolltop?action=raw&ctype=text/css"> | <link rel="stylesheet" href="https://2018.igem.org/Team:XMU-China/css/material-scrolltop?action=raw&ctype=text/css"> | ||
| Line 35: | Line 35: | ||
<body> | <body> | ||
| − | < | + | <header></header> |
| + | <div id="container"> | ||
<header> | <header> | ||
<div class="wrapper cf"> | <div class="wrapper cf"> | ||
| Line 45: | Line 46: | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Description">Description</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Description">Description</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Design">Design</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Design">Design</a></li> | ||
| − | |||
<li><a href="https://2018.igem.org/Team:XMU-China/Results">Results</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Results">Results</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Demonstrate">Demonstrate</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Parts">Parts</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Parts">Parts</a></li> | ||
</ul> | </ul> | ||
| Line 56: | Line 57: | ||
<ul> | <ul> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Hardware">Overview</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware">Overview</a></li> | ||
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Microfluidic_Chips">Microfluidic Chips</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Fluorescence_Detection">Fluorescence Detection</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Raspberry_Pi">Raspberry Pi</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/ | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Application">Application</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Software"> | + | <li><a href="https://2018.igem.org/Team:XMU-China/Software">Software</a></li> |
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Applied_Design">Product Design</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 66: | Line 68: | ||
<a href="#">Model</a> | <a href="#">Model</a> | ||
<ul> | <ul> | ||
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Model"> | + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Summary">Summary</a></li> |
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Thermodynamic_model">Thermodynamic Model</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Fluid_dynamics_model">Fluid dynamics Model</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Molecular_docking_model">Molecular Docking Model</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#The_dynamic_model">Derivation of Rate Equation</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
<li class="Human_Practice"> | <li class="Human_Practice"> | ||
| − | <a href="#"> | + | <a href="#">Social Works</a> |
<ul> | <ul> | ||
| − | <li><a href="https://2018.igem.org/ | + | <li><a href="https://2018.igem.org/Team:XMU-China/Human_Practices">Human Practice</a></li> |
| − | + | ||
| − | + | ||
| − | + | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Public_Engagement">Engagement</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Public_Engagement">Engagement</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Collaborations">Collaborations</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Collaborations">Collaborations</a></li> | ||
| Line 95: | Line 98: | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Notebook">Notebook</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Notebook">Notebook</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Experiments">Experiments</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Experiments">Experiments</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Engineering">Engineering</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 103: | Line 107: | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Attributions">Attributions</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Attributions">Attributions</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Judging">Judging</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Judging">Judging</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/After_iGEM">After iGEM</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 110: | Line 115: | ||
</div> | </div> | ||
</header> | </header> | ||
| − | </div | + | </div> |
<script src="js/jquery-1.11.0.min.js"></script> | <script src="js/jquery-1.11.0.min.js"></script> | ||
<!-- <script src="js/hc-mobile-nav.js"></script> --> | <!-- <script src="js/hc-mobile-nav.js"></script> --> | ||
| Line 120: | Line 125: | ||
</div> | </div> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
| − | + | <div class="nav"> | |
<div id="Team"> | <div id="Team"> | ||
<div class="nav-word">Team</div> | <div class="nav-word">Team</div> | ||
| Line 127: | Line 132: | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Attributions">Attributions</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Attributions">Attributions</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Judging">Judging</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Judging">Judging</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/After_iGEM">After iGEM</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
<div id="Notebook"> | <div id="Notebook"> | ||
| − | + | <div class="nav-word">Notebook</div> | |
| − | + | ||
| − | + | ||
<ul> | <ul> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Notebook">Notebook</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Notebook">Notebook</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Experiments">Experiments</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Experiments">Experiments</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Engineering">Engineering</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| Line 148: | Line 153: | ||
</div> | </div> | ||
<div id="Human_Practice"> | <div id="Human_Practice"> | ||
| − | <div class="nav-word"> | + | <div class="nav-word">Social Works</div> |
<ul> | <ul> | ||
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Human_Practices"> | + | <li><a href="https://2018.igem.org/Team:XMU-China/Human_Practices">Human Practice</a></li> |
| − | + | ||
| − | + | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Public_Engagement">Engagement</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Public_Engagement">Engagement</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Collaborations">Collaborations</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Collaborations">Collaborations</a></li> | ||
| Line 161: | Line 164: | ||
<div class="nav-word">Model</div> | <div class="nav-word">Model</div> | ||
<ul> | <ul> | ||
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Model"> | + | <li><a href="https://2018.igem.org/Team:XMU-China/Model">Summary</a></li> |
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Thermodynamic_model">Thermodynamic Model</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Fluid_dynamics_model">Fluid dynamics Model</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#Molecular_docking_model">Molecular Docking Model</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Model#The_dynamic_model">Derivation of Rate Equation</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| Line 168: | Line 175: | ||
<ul> | <ul> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Hardware">Overview</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware">Overview</a></li> | ||
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Microfluidic_Chips">Microfluidic Chips</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Fluorescence_Detection">Fluorescence Detection</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Raspberry_Pi">Raspberry Pi</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/ | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware#Application">Application</a></li> |
| − | <li><a href="https://2018.igem.org/Team:XMU-China/Software"> | + | <li><a href="https://2018.igem.org/Team:XMU-China/Software">Software</a></li> |
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Applied_Design">Product Design</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| Line 180: | Line 188: | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Description">Description</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Description">Description</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Design">Design</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Design">Design</a></li> | ||
| − | |||
<li><a href="https://2018.igem.org/Team:XMU-China/Results">Results</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Results">Results</a></li> | ||
| + | <li><a href="https://2018.igem.org/Team:XMU-China/Demonstrate">Demonstrate</a></li> | ||
<li><a href="https://2018.igem.org/Team:XMU-China/Parts">Parts</a></li> | <li><a href="https://2018.igem.org/Team:XMU-China/Parts">Parts</a></li> | ||
</ul> | </ul> | ||
| Line 190: | Line 198: | ||
</a> | </a> | ||
</div> | </div> | ||
| − | </div | + | </div> |
</div> | </div> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
| Line 201: | Line 209: | ||
<a href="#ABCDsystem" class="Quick-navigation-item"> | <a href="#ABCDsystem" class="Quick-navigation-item"> | ||
<img id="turn_img" src="https://static.igem.org/mediawiki/2018/d/db/T--XMU-China--right1.png"> | <img id="turn_img" src="https://static.igem.org/mediawiki/2018/d/db/T--XMU-China--right1.png"> | ||
| − | <a href="#ABCDsystem"id="Quick_A"> | + | <a href="#ABCDsystem"id="Quick_A">ABCD system</a></a> |
<a href="#OMVs" class="Quick-navigation-item" > | <a href="#OMVs" class="Quick-navigation-item" > | ||
<img id="turn_img" src="https://static.igem.org/mediawiki/2018/c/cd/T--XMU-China--right2.png"> | <img id="turn_img" src="https://static.igem.org/mediawiki/2018/c/cd/T--XMU-China--right2.png"> | ||
| Line 213: | Line 221: | ||
<section id="ABCDsystem" class="js-scroll-step"> | <section id="ABCDsystem" class="js-scroll-step"> | ||
<div class="headline"> | <div class="headline"> | ||
| − | + | ABCD system | |
</div> | </div> | ||
<h1>Ideas</h1> | <h1>Ideas</h1> | ||
| − | <p>During our brainstorming, we read the project of the 2017 Munich team, which established a system for the universal detection of RNA biomarkers. Therefore, we hope to improve the entire detection system and establish a system that can detect protein biomarkers universally. < | + | <p>During our brainstorming, we read the project of the <a class="click_here" href="https://2017.igem.org/Team:Munich">2017 Munich team</a>, which established a system for the universal detection of RNA biomarkers. Therefore, we hope to improve the entire detection system and establish a system that can detect protein biomarkers universally. </p> |
| − | + | <p>Luckily, our ABCD system has appeared. Later, we discussed how could it be implied in synthetic biology. We found the answer ultimately in cell-free synthetic biology. In addition, after exchanging ideas with the Munich team, we were more certain that we did use engineering principles to re-edit the biological system.</p> <p>We have divided our ABCD system into three modules, using engineering ideas, so that each can be "plug and play", but also can be "replaced". The three modules are "Sensor", "Amplifier" and "Reporter".</p> | |
| + | |||
<p class="F1"> | <p class="F1"> | ||
<img src="https://static.igem.org/mediawiki/2018/4/45/T--XMU-China--ABCD_design1.png"> | <img src="https://static.igem.org/mediawiki/2018/4/45/T--XMU-China--ABCD_design1.png"> | ||
| + | <p class="Figure_word"><strong>Figure 1.</strong> The whole design.</p> | ||
</p> | </p> | ||
<h1>Sensor</h1> | <h1>Sensor</h1> | ||
<p>In the Sensor, we first selected the epithelial cell adhesion molecule as the object of our detection. The epithelial cell adhesion molecule (EpCAM, also known as ESA or CD326) is a transmembrane glycoprotein that mediates epithelial cell-specific intercellular adhesion and involves cell signaling, migration, proliferation, and differentiation. EpCAM has been detected in most adenocarcinomas, and is also found in metastatic tumors, malignant effusions and cancer stem cells. | <p>In the Sensor, we first selected the epithelial cell adhesion molecule as the object of our detection. The epithelial cell adhesion molecule (EpCAM, also known as ESA or CD326) is a transmembrane glycoprotein that mediates epithelial cell-specific intercellular adhesion and involves cell signaling, migration, proliferation, and differentiation. EpCAM has been detected in most adenocarcinomas, and is also found in metastatic tumors, malignant effusions and cancer stem cells. | ||
</p> | </p> | ||
| − | <p>Next, we | + | <p>Next, we choosed the ssDNA aptamer as an intermediary for downstream signaling of proteins. It is a single-stranded DNA that specifically binds to a target molecule. Theoretically, the corresponding aptamer of any protein can be screened by SELEX technique. And we choosed EpCAM's aptamer--SYL3C. <sup>[1]</sup></p> |
| − | < | + | <div class="table_container"> |
| − | + | <table class="table"> | |
<tr> | <tr> | ||
<th>Name</th> | <th>Name</th> | ||
| Line 245: | Line 255: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| − | <p> At the same time, we also | + | </div> |
| + | <p> At the same time, we also used the competition method to facilitate our design of hardware. As shown in the figure, according to our assumption, the upstream EpCAM would compete with our designed aptamer and its partial complementary chain. After that, some complementary strands would be competed and freed, coming out and going to the next module to get the signal amplified. | ||
</p> | </p> | ||
<p class="F1"> | <p class="F1"> | ||
<img src="https://static.igem.org/mediawiki/2018/d/d5/T--XMU-China--ABCD_design2.png"> | <img src="https://static.igem.org/mediawiki/2018/d/d5/T--XMU-China--ABCD_design2.png"> | ||
| − | <p class="Figure_word"> | + | <p class="Figure_word"><strong>Figure 2.</strong> The principle of Sensor.</p> |
</p> | </p> | ||
<p> | <p> | ||
| Line 255: | Line 266: | ||
</p> | </p> | ||
<h1>Amplifier</h1> | <h1>Amplifier</h1> | ||
| − | <p> | + | <p>In this module we used the Cas12a protein to amplify the signal. Cas12a (also known as Cpf1) was originally discovered as a replacement for Cas9. <sup>[2]</sup> But this year's discovery showed that it had a new function of being activated by ssDNA and realizing trans-cleavage. <sup>[3]</sup> |
| − | + | ||
<p class="F1"> | <p class="F1"> | ||
<img src="https://static.igem.org/mediawiki/2018/9/90/T--XMU-China--ABCD_design3.png"> | <img src="https://static.igem.org/mediawiki/2018/9/90/T--XMU-China--ABCD_design3.png"> | ||
| − | + | <P class="Figure_word"><strong>Figure 3.</strong> The principle of Amplifer.</P> | |
| − | < | + | </p> |
| + | <p>As shown in the figure, when some of our upstream complementary strands are released to the Cas12a protein after competitions, recognized by the specifically designed crRNA and finally complemented with it successfully, the Cas12a protein will be activated, and non-specific cleavage will occur.</p> | ||
<table class="table"> | <table class="table"> | ||
<tr> | <tr> | ||
| Line 275: | Line 286: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| − | <h1> | + | <h1>Reporter</h1> |
<p>When the Cas12a protein is activated, it will realize a non-specific cleavage that functions as DNase I, so we can report our signal by using the method of detecting DNase I.</p> | <p>When the Cas12a protein is activated, it will realize a non-specific cleavage that functions as DNase I, so we can report our signal by using the method of detecting DNase I.</p> | ||
| − | <p>We finally used the fluorescent signal as our reporting signal, because it was easy to observe, semi-quantitative, and can be directly read and analyzed by the machine. And our signal reports used the commercially available DNaseAlert kit on the market. <sup>[4]</sup></p> | + | <p>We finally used the fluorescent signal as our reporting signal, because it was easy to observe, semi-quantitative, and can be directly read and analyzed by the machine. And our signal reports used the commercially available DNaseAlert<sup>TM</sup> kit on the market. <sup>[4]</sup></p> |
| − | <p>The working principle is as follows: a combination of a ground state fluorescent molecule and a quencher generates a complex by ssDNA, and the complex quenches the fluorescence. When the nuclease | + | <p>The working principle is as follows: a combination of a ground state fluorescent molecule and a quencher generates a complex by ssDNA, and the complex quenches the fluorescence. When the nuclease cleavages the ssDNA, the quenching fails and the fluorophore re-emits.</p> |
| − | + | ||
<p class="F1"> | <p class="F1"> | ||
<img src="https://static.igem.org/mediawiki/2018/3/31/T--XMU-China--ABCD_design4.png"> | <img src="https://static.igem.org/mediawiki/2018/3/31/T--XMU-China--ABCD_design4.png"> | ||
| + | <p class="Figure_word"><strong>Figure 4.</strong> The principle of Reporter.</p> | ||
</p> | </p> | ||
| − | + | <h1>References</h1> | |
| − | + | <p class="reference"> | |
| − | < | + | [1] Yanling Song, Zhi Zhu, Yuan An, Weiting Zhang, Huimin Zhang, Dan Liu, Chundong Yu, Wei Duan, Chaoyong James Yang. Selection of DNA Aptamers against Epithelial Cell Adhesion Molecule for Cancer Cell Imaging and Circulating Tumor Cell Capture. <i>Anal. Chem</i>. <strong>2013</strong>, 85: 4141-4149. <br> |
| − | + | [2] Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. <i>Cell</i>. <strong>2015</strong>, 163(3): 759-771. <br> | |
| − | <p | + | [3] Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. <i>Science</i>. <strong>2018</strong>, 360(6387): eaar6245. <br> |
| − | + | [4] <a class="click_here" href="https://static.igem.org/mediawiki/2018/4/4c/T--XMU-China--dnasealert-detection-manual.pdf">DNaseAlert<sup>TM</sup> Substrate Nuclease Detection System User Manual.</a> <br> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | [1] Yanling Song, Zhi Zhu, Yuan An, Weiting Zhang, Huimin Zhang, Dan Liu, Chundong Yu, Wei Duan, Chaoyong James Yang | + | |
| − | [2] Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F | + | |
| − | [3] Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA | + | |
| − | [4] | + | |
| − | + | ||
| − | + | ||
</p> | </p> | ||
</section> | </section> | ||
| Line 308: | Line 307: | ||
</div> | </div> | ||
<h1>Ideas</h1> | <h1>Ideas</h1> | ||
| − | <p>To utilize outer-membrane vesicles (OMVs) as our cell-free platform for RNA delivery and then for disease treatment, we | + | <p>To utilize outer-membrane vesicles (OMVs) as our cell-free platform for RNA delivery and then for disease treatment, we designed three modules with different functions, which can be divided into OmpA, L7Ae, and Box (Figure 1). |
</p> | </p> | ||
<p class="F2"> | <p class="F2"> | ||
<img src="https://static.igem.org/mediawiki/2018/b/b6/T--XMU-China--OMVs21.png"> | <img src="https://static.igem.org/mediawiki/2018/b/b6/T--XMU-China--OMVs21.png"> | ||
| − | <p class="Figure_word">Figure 1. a) Genetic circuit of three modules. b) Schematic illustration of our total circuit.</p> | + | <p class="Figure_word"><strong>Figure 1.</strong> a) Genetic circuit of three modules. b) Schematic illustration of our total circuit.</p> |
</p> | </p> | ||
<h1>Module one - OmpA</h1> | <h1>Module one - OmpA</h1> | ||
| − | <p>Porin outer-membrane protein A, OmpA, is a kind of highly expressed transmembrane porin protein of E. coli and is therefore abundant in OMVs <sup>[1]</sup> | + | <p>Porin outer-membrane protein A, OmpA, is a kind of highly expressed transmembrane porin protein of <i>E. coli</i> and is therefore abundant in OMVs. <sup>[1]</sup> We inserted SpyTag to the C-termini and N-termini of a truncated form of OmpA respectively to compare the efficiency of SpyTag/SpyCatcher linkage (Figure 2). </p> |
<p class="F2"> | <p class="F2"> | ||
<img src="https://static.igem.org/mediawiki/2018/c/ce/T--XMU-China--OMVs22.png"> | <img src="https://static.igem.org/mediawiki/2018/c/ce/T--XMU-China--OMVs22.png"> | ||
| − | <p class="Figure_word">Figure 2. a) Genetic circuit of SpyTag-OmpA part and OmpA-SpyTag part. b) Schematic illustration of module one.</p> | + | <p class="Figure_word"><strong>Figure 2.</strong> a) Genetic circuit of SpyTag-OmpA part and OmpA-SpyTag part. b) Schematic illustration of module one.</p> |
</p> | </p> | ||
| − | <p>Furthermore, to characterize whether our OmpA-SpyTag can localize in OMVs, we also | + | <p>Furthermore, to characterize whether our OmpA-SpyTag can localize in OMVs, we also attached GFP to both OmpA parts (Figure 3). </p> |
<p class="F2"> | <p class="F2"> | ||
<img src="https://static.igem.org/mediawiki/2018/d/db/T--XMU-China--OMVs23.png"> | <img src="https://static.igem.org/mediawiki/2018/d/db/T--XMU-China--OMVs23.png"> | ||
| − | <p class="Figure_word">Figure 3. a) Genetic circuit of SpyTag-OmpA-GFP part and OmpA-SpyTag-GFP part. b) Schematic illustration of these two parts.</p> | + | <p class="Figure_word"><strong>Figure 3.</strong> a) Genetic circuit of SpyTag-OmpA-GFP part and OmpA-SpyTag-GFP part. b) Schematic illustration of these two parts.</p> |
</p> | </p> | ||
<h1>Module two – L7Ae</h1> | <h1>Module two – L7Ae</h1> | ||
| − | <p>Archaeal ribosomal protein protein L7Ae owns the ability to bind with RNA with C/ | + | <p>Archaeal ribosomal protein protein L7Ae owns the ability to bind with RNA with C/D<sub>box</sub> RNA structure. <sup>[2-4]</sup> Through the isopeptide bonds formed between SpyTag and SpyCatcher, L7Ae can be anchored with OmpA, and then secreted into OMVs. To transport our L7Ae into the periplasmic space, we inserted TorA leader sequence to the circuit (Figure 4). </p> |
<p class="F2"><img src="https://static.igem.org/mediawiki/2018/c/c3/T--XMU-China--OMVs24.png"> | <p class="F2"><img src="https://static.igem.org/mediawiki/2018/c/c3/T--XMU-China--OMVs24.png"> | ||
| − | <p class="Figure_word">Figure 4. Genetic circuit of SpyCatcher-L7Ae.</p> | + | <p class="Figure_word"><strong>Figure 4.</strong> Genetic circuit of SpyCatcher-L7Ae.</p> |
</p> | </p> | ||
| − | <p>To localize our L7Ae inside OMVs, we also | + | <p>To localize our L7Ae inside OMVs, we also conjugated mRFP1 to the L7Ae-SpyCatcher. Under the induction of the IPTG, our bacteria coudld express our L7Ae parts (Figure 5). </p> |
<p class="F2"><img src="https://static.igem.org/mediawiki/2018/3/37/T--XMU-China--OMVs25.png"> | <p class="F2"><img src="https://static.igem.org/mediawiki/2018/3/37/T--XMU-China--OMVs25.png"> | ||
| − | <p class="Figure_word">Figure 5. Genetic circuit of SpyCatcher-L7Ae-RFP.</p> | + | <p class="Figure_word"><strong>Figure 5.</strong> Genetic circuit of SpyCatcher-L7Ae-RFP.</p> |
</p> | </p> | ||
<h1>Module three - Box</h1> | <h1>Module three - Box</h1> | ||
| − | <p>We inserted a C/ | + | <p>We inserted a C/D<sub>box</sub> into the 3'-untranslated region (3'-UTR) of the gene coding for siRNA. <sup>[5]</sup> Under the induction of 0.2% Arabinose, our bacteria could express our Box parts (Figure 6). </p> |
<p class="F2"><img src="https://static.igem.org/mediawiki/2018/d/db/T--XMU-China--OMVs26.png"> | <p class="F2"><img src="https://static.igem.org/mediawiki/2018/d/db/T--XMU-China--OMVs26.png"> | ||
| − | <p class="Figure_word">Figure 6. Genetic circuit of Box part.</p> | + | <p class="Figure_word"><strong>Figure 6.</strong> Genetic circuit of Box part.</p> |
</p> | </p> | ||
| − | <p>Under the induction of 0.2% Arabinose and then 0.5 mM IPTG, we | + | <p>Under the induction of 0.2% Arabinose and then 0.5 mM IPTG, we could produce L7Ae-SpyCatcher and siRNA-C/D<sub>box</sub>. With the isopeptide bond formed between SpyTag and SpyCatcher, and the ability of L7Ae to be bind with C/D<sub>box</sub>, we could produce customizable and cell-free OMVs containing specific siRNA to target for oncogenic gene (Figure 7). </p> |
<p class="F2"><img src="https://static.igem.org/mediawiki/2018/a/ac/T--XMU-China--OMVs27.png"> | <p class="F2"><img src="https://static.igem.org/mediawiki/2018/a/ac/T--XMU-China--OMVs27.png"> | ||
| − | <p class="Figure_word">Figure | + | <p class="Figure_word"><strong>Figure 7.</strong> Schematic illustration of our total circuit.</p> |
</p> | </p> | ||
| − | <h1 class=" | + | <h1 class="references">References</h1> |
| − | <p> | + | <p class="reference"> |
| − | [1] Acevedo R, Fernández S, Zayas C, et al. Bacterial Outer Membrane Vesicles and Vaccine Applications | + | [1] Acevedo R, Fernández S, Zayas C, <i>et al</i>. Bacterial Outer Membrane Vesicles and Vaccine Applications. <i>Frontiers in Immunology</i>. <strong>2014</strong>, 5: 121. <br> |
| − | [2] | + | [2] Ausländer S, Ausländer D, Müller M, <i>et al</i>. Programmable single-cell mammalian biocomputers. <i>Nature</i>, <strong>2012</strong>, 487(7405):123-127. <br> |
| − | [3] Saito H, Fujita Y, Kashida S, et al. Synthetic human cell fate regulation by protein-driven RNA switches | + | [3] Saito H, Fujita Y, Kashida S, <i>et al</i>. Synthetic human cell fate regulation by protein-driven RNA switches. <i>Nature Communications</i>. <strong>2011</strong>, 2(1): 160. <br> |
| − | [4] Saito H, Kobayashi T, Hara T, et al. Synthetic translational regulation by an L7Ae–kink-turn RNP switch | + | [4] Saito H, Kobayashi T, Hara T, <i>et al</i>. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. <i>Nature Chemical Biology</i>. <strong>2009</strong>, 6(1): 71. <br> |
| − | [5] Kojima R, Bojar D, Rizzi G, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for | + | [5] Kojima R, Bojar D, Rizzi G, <i>et al</i>. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. <i>Nature Communications</i>. <strong>2018</strong>, 9(1): 1305. <br> |
</p> | </p> | ||
</section> | </section> | ||
| Line 357: | Line 356: | ||
</div> | </div> | ||
<h1>Ideas</h1> | <h1>Ideas</h1> | ||
| − | <p>We selected two proteins SAHS1 and CAHS1, which | + | <p>We selected two proteins SAHS1 and CAHS1, which have been shown with excellent effects by a Japanese team in 2012. SAHS1 could be used to preserve proteins and CAHS1 could be used to preserve bacteria. |
</p> | </p> | ||
| − | + | ||
| − | <p> | + | <p>Our genetic circuits are shown below:</p> |
<p class="F2"> | <p class="F2"> | ||
<img src="https://static.igem.org/mediawiki/2018/e/e6/T--XMU-China--TDP3.png"> | <img src="https://static.igem.org/mediawiki/2018/e/e6/T--XMU-China--TDP3.png"> | ||
| − | <p class="Figure_word"> | + | <p class="Figure_word"><strong>Figure1.</strong> Genetic circuits of TDP.</p> |
</p> | </p> | ||
| − | <p>CAHS is a cytoplasmic-abundant protein which can | + | <p>CAHS protein is a cytoplasmic-abundant protein which can strengthen the stress tolerance of bacteria. Taking biosafety into account, we added constant promoter J23100 into the CAHS circuit to express lacI in order to inhibit lac O promoter. Once IPTG is added, the inhibition of lacI will be removed. Then CAHS and mRFP will be expressed, in which mRFP was used for screening. We intended to adopt the Red recombination method to transform the circuit into the genome of DH5α. Hence, DH5α would be improved and preserved easily.</p> |
| − | <p> | + | <p>After discussion, we decided to add mRFP or blue chromoprotein to the SAHS part for selecting <i>E. coli</i> transfected successfully. Since SAHS protein is an exogenous protein, it is preceded by a signal peptide, which allows it to pass through the cell membrane. We removed the LB medium by centrifugation and harvested our proteins in high concentration. As a protective agent, SAHS can prevent Cas12a from lyophilization.</p> |
<h1>Expectation</h1> | <h1>Expectation</h1> | ||
<h2>A new E.coli strain</h2> | <h2>A new E.coli strain</h2> | ||
| − | <p> We intend to transform the circuit into the genome of DH5α, | + | <p> We intend to transform the circuit into the genome of DH5α. Hence, DH5α would be improved and preserved easily. In this way, <i>E. coli</i> DH5α can be transported normally and easily. |
</p> | </p> | ||
<h2>A new way to preserve protein</h2> | <h2>A new way to preserve protein</h2> | ||
| − | <p> We hope that the purified TDPs can | + | <p> We hope that the purified TDPs can be used to preserve proteins and other biological materials. </p> |
| − | + | ||
| − | + | ||
| − | + | ||
<h1>References</h1> | <h1>References</h1> | ||
<p class="reference"> | <p class="reference"> | ||
| − | [1] | + | [1] Yamaguchi A, Tanaka S, Yamaguchi S, <i>et al</i>. Two Novel Heat-Soluble Protein Families Abundantly Expressed in an Anhydrobiotic Tardigrade. <i>PLoS ONE</i>, <strong>2012</strong>, 7(8):e44209. <br> |
| − | [2] | + | [2] Boothby T C, Tapia H, Brozena A H, <i>et al</i>. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. <i>Molecular Cell</i>, <strong>2017</strong>, 65(6):975-984. <br> |
| − | [3] | + | [3] Boothby T C, Pielak G J. Intrinsically Disordered Proteins and Desiccation Tolerance: Elucidating Functional and Mechanistic Underpinnings of Anhydrobiosis. <i>Bioessays</i>, <strong>2017</strong>, 39(11). <br> |
| − | + | ||
</p> | </p> | ||
</section> | </section> | ||
| Line 396: | Line 391: | ||
<div class="footer"> | <div class="footer"> | ||
<div class="footer_top"> | <div class="footer_top"> | ||
| − | + | <img src="https://static.igem.org/mediawiki/2018/3/3d/T--XMU-China--xmu_is_different.png"> | |
| − | + | <ul> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China">Home</a></a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Model">Model</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Parts">Parts</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Entrepreneurship">Entrepreneurship</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Attributions">Attributions</a></li> | |
| − | + | </ul> | |
| − | + | <ul> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Design">Design</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Hardware">Hardware</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Human_Practices">Human Practices</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Notebook">Notebook</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Judging">Judging</a></li> | |
| − | + | </ul> | |
| − | + | <ul> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Results">Results</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Software">Software</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Collaborations">Collaborations</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/Members">Members</a></li> | |
| − | + | <li><a href="https://2018.igem.org/Team:XMU-China/After_iGEM">After iGEM</a></li> | |
| − | + | </ul> | |
| − | + | </div> | |
| − | + | ||
</div> | </div> | ||
<div class="bottom"></div> | <div class="bottom"></div> | ||
Latest revision as of 03:49, 18 October 2018


Ideas
During our brainstorming, we read the project of the 2017 Munich team, which established a system for the universal detection of RNA biomarkers. Therefore, we hope to improve the entire detection system and establish a system that can detect protein biomarkers universally.
Luckily, our ABCD system has appeared. Later, we discussed how could it be implied in synthetic biology. We found the answer ultimately in cell-free synthetic biology. In addition, after exchanging ideas with the Munich team, we were more certain that we did use engineering principles to re-edit the biological system.
We have divided our ABCD system into three modules, using engineering ideas, so that each can be "plug and play", but also can be "replaced". The three modules are "Sensor", "Amplifier" and "Reporter".

Figure 1. The whole design.
Sensor
In the Sensor, we first selected the epithelial cell adhesion molecule as the object of our detection. The epithelial cell adhesion molecule (EpCAM, also known as ESA or CD326) is a transmembrane glycoprotein that mediates epithelial cell-specific intercellular adhesion and involves cell signaling, migration, proliferation, and differentiation. EpCAM has been detected in most adenocarcinomas, and is also found in metastatic tumors, malignant effusions and cancer stem cells.
Next, we choosed the ssDNA aptamer as an intermediary for downstream signaling of proteins. It is a single-stranded DNA that specifically binds to a target molecule. Theoretically, the corresponding aptamer of any protein can be screened by SELEX technique. And we choosed EpCAM's aptamer--SYL3C. [1]
| Name | Sequence | Modification |
|---|---|---|
| SYL3C | 5'-CACTACAGAGGTTGCGTCTGTCCC ACGTTGTCATGGGGGGTTGGCCTG-3' | 3'-Biotin-TEG |
| Complementary strand | 5'-CTCTGTAGTGTTTTTTTTTTTTTT-3' | 3'-FITC |
At the same time, we also used the competition method to facilitate our design of hardware. As shown in the figure, according to our assumption, the upstream EpCAM would compete with our designed aptamer and its partial complementary chain. After that, some complementary strands would be competed and freed, coming out and going to the next module to get the signal amplified.

Figure 2. The principle of Sensor.
For this module, we also modeled the molecular docking model of aptamer and protein, and partially predicted the number of bases in the complementary strands. Click to learn more.
Amplifier
In this module we used the Cas12a protein to amplify the signal. Cas12a (also known as Cpf1) was originally discovered as a replacement for Cas9. [2] But this year's discovery showed that it had a new function of being activated by ssDNA and realizing trans-cleavage. [3]

Figure 3. The principle of Amplifer.
As shown in the figure, when some of our upstream complementary strands are released to the Cas12a protein after competitions, recognized by the specifically designed crRNA and finally complemented with it successfully, the Cas12a protein will be activated, and non-specific cleavage will occur.
| Name | Sequence | Modification |
|---|---|---|
| SYL3C-crRNA | 5'-UAAUUUCUACUCUUGUAGAU AAAAAAAAAAACACUACAGAG-3 | None |
Reporter
When the Cas12a protein is activated, it will realize a non-specific cleavage that functions as DNase I, so we can report our signal by using the method of detecting DNase I.
We finally used the fluorescent signal as our reporting signal, because it was easy to observe, semi-quantitative, and can be directly read and analyzed by the machine. And our signal reports used the commercially available DNaseAlertTM kit on the market. [4]
The working principle is as follows: a combination of a ground state fluorescent molecule and a quencher generates a complex by ssDNA, and the complex quenches the fluorescence. When the nuclease cleavages the ssDNA, the quenching fails and the fluorophore re-emits.

Figure 4. The principle of Reporter.
References
[1] Yanling Song, Zhi Zhu, Yuan An, Weiting Zhang, Huimin Zhang, Dan Liu, Chundong Yu, Wei Duan, Chaoyong James Yang. Selection of DNA Aptamers against Epithelial Cell Adhesion Molecule for Cancer Cell Imaging and Circulating Tumor Cell Capture. Anal. Chem. 2013, 85: 4141-4149.
[2] Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015, 163(3): 759-771.
[3] Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018, 360(6387): eaar6245.
[4] DNaseAlertTM Substrate Nuclease Detection System User Manual.
Ideas
To utilize outer-membrane vesicles (OMVs) as our cell-free platform for RNA delivery and then for disease treatment, we designed three modules with different functions, which can be divided into OmpA, L7Ae, and Box (Figure 1).

Figure 1. a) Genetic circuit of three modules. b) Schematic illustration of our total circuit.
Module one - OmpA
Porin outer-membrane protein A, OmpA, is a kind of highly expressed transmembrane porin protein of E. coli and is therefore abundant in OMVs. [1] We inserted SpyTag to the C-termini and N-termini of a truncated form of OmpA respectively to compare the efficiency of SpyTag/SpyCatcher linkage (Figure 2).

Figure 2. a) Genetic circuit of SpyTag-OmpA part and OmpA-SpyTag part. b) Schematic illustration of module one.
Furthermore, to characterize whether our OmpA-SpyTag can localize in OMVs, we also attached GFP to both OmpA parts (Figure 3).

Figure 3. a) Genetic circuit of SpyTag-OmpA-GFP part and OmpA-SpyTag-GFP part. b) Schematic illustration of these two parts.
Module two – L7Ae
Archaeal ribosomal protein protein L7Ae owns the ability to bind with RNA with C/Dbox RNA structure. [2-4] Through the isopeptide bonds formed between SpyTag and SpyCatcher, L7Ae can be anchored with OmpA, and then secreted into OMVs. To transport our L7Ae into the periplasmic space, we inserted TorA leader sequence to the circuit (Figure 4).

Figure 4. Genetic circuit of SpyCatcher-L7Ae.
To localize our L7Ae inside OMVs, we also conjugated mRFP1 to the L7Ae-SpyCatcher. Under the induction of the IPTG, our bacteria coudld express our L7Ae parts (Figure 5).

Figure 5. Genetic circuit of SpyCatcher-L7Ae-RFP.
Module three - Box
We inserted a C/Dbox into the 3'-untranslated region (3'-UTR) of the gene coding for siRNA. [5] Under the induction of 0.2% Arabinose, our bacteria could express our Box parts (Figure 6).

Figure 6. Genetic circuit of Box part.
Under the induction of 0.2% Arabinose and then 0.5 mM IPTG, we could produce L7Ae-SpyCatcher and siRNA-C/Dbox. With the isopeptide bond formed between SpyTag and SpyCatcher, and the ability of L7Ae to be bind with C/Dbox, we could produce customizable and cell-free OMVs containing specific siRNA to target for oncogenic gene (Figure 7).
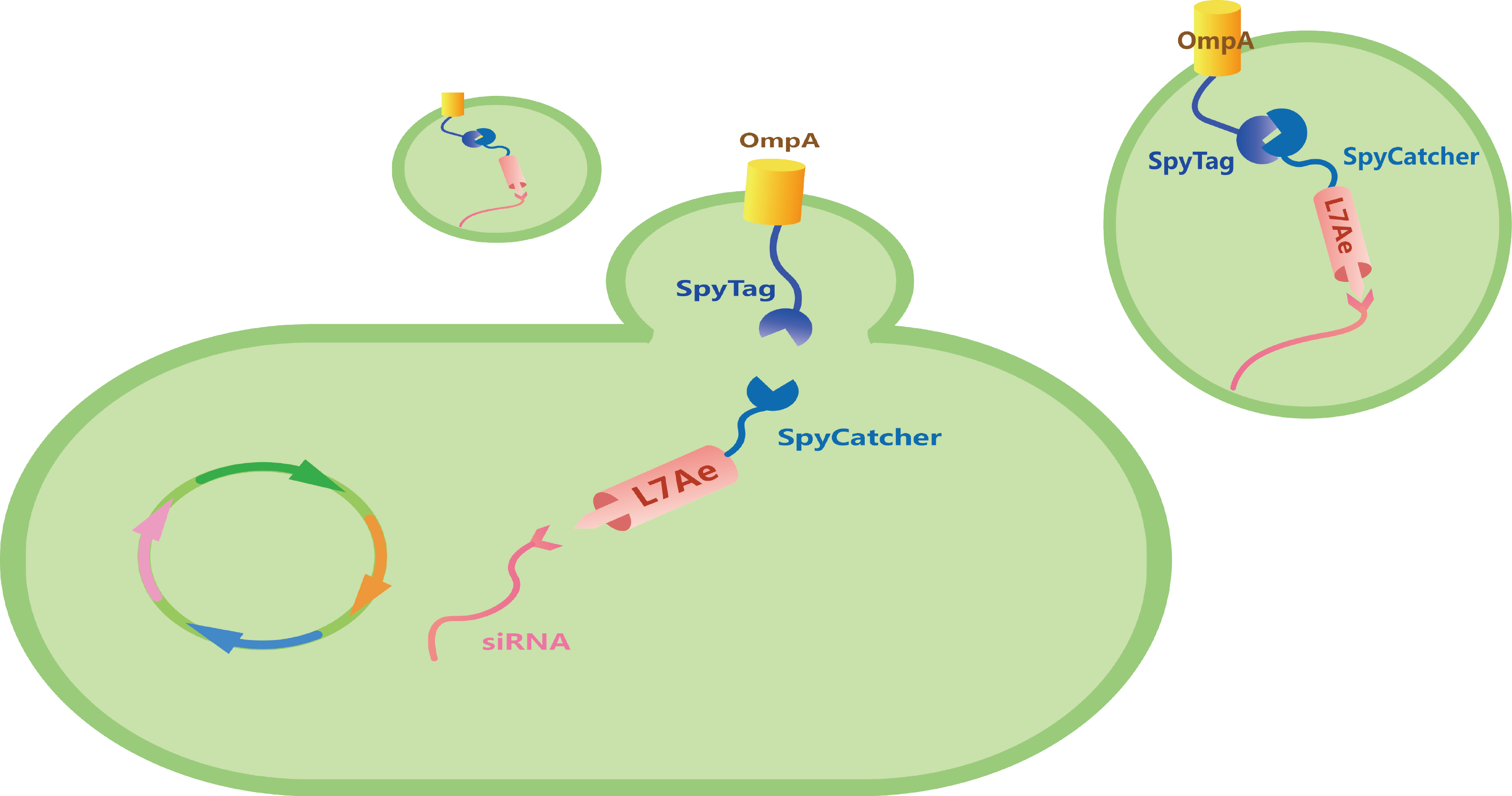
Figure 7. Schematic illustration of our total circuit.
References
[1] Acevedo R, Fernández S, Zayas C, et al. Bacterial Outer Membrane Vesicles and Vaccine Applications. Frontiers in Immunology. 2014, 5: 121.
[2] Ausländer S, Ausländer D, Müller M, et al. Programmable single-cell mammalian biocomputers. Nature, 2012, 487(7405):123-127.
[3] Saito H, Fujita Y, Kashida S, et al. Synthetic human cell fate regulation by protein-driven RNA switches. Nature Communications. 2011, 2(1): 160.
[4] Saito H, Kobayashi T, Hara T, et al. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. Nature Chemical Biology. 2009, 6(1): 71.
[5] Kojima R, Bojar D, Rizzi G, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nature Communications. 2018, 9(1): 1305.
Ideas
We selected two proteins SAHS1 and CAHS1, which have been shown with excellent effects by a Japanese team in 2012. SAHS1 could be used to preserve proteins and CAHS1 could be used to preserve bacteria.
Our genetic circuits are shown below:

Figure1. Genetic circuits of TDP.
CAHS protein is a cytoplasmic-abundant protein which can strengthen the stress tolerance of bacteria. Taking biosafety into account, we added constant promoter J23100 into the CAHS circuit to express lacI in order to inhibit lac O promoter. Once IPTG is added, the inhibition of lacI will be removed. Then CAHS and mRFP will be expressed, in which mRFP was used for screening. We intended to adopt the Red recombination method to transform the circuit into the genome of DH5α. Hence, DH5α would be improved and preserved easily.
After discussion, we decided to add mRFP or blue chromoprotein to the SAHS part for selecting E. coli transfected successfully. Since SAHS protein is an exogenous protein, it is preceded by a signal peptide, which allows it to pass through the cell membrane. We removed the LB medium by centrifugation and harvested our proteins in high concentration. As a protective agent, SAHS can prevent Cas12a from lyophilization.
Expectation
A new E.coli strain
We intend to transform the circuit into the genome of DH5α. Hence, DH5α would be improved and preserved easily. In this way, E. coli DH5α can be transported normally and easily.
A new way to preserve protein
We hope that the purified TDPs can be used to preserve proteins and other biological materials.
References
[1] Yamaguchi A, Tanaka S, Yamaguchi S, et al. Two Novel Heat-Soluble Protein Families Abundantly Expressed in an Anhydrobiotic Tardigrade. PLoS ONE, 2012, 7(8):e44209.
[2] Boothby T C, Tapia H, Brozena A H, et al. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Molecular Cell, 2017, 65(6):975-984.
[3] Boothby T C, Pielak G J. Intrinsically Disordered Proteins and Desiccation Tolerance: Elucidating Functional and Mechanistic Underpinnings of Anhydrobiosis. Bioessays, 2017, 39(11).


