| (43 intermediate revisions by 6 users not shown) | |||
| Line 6: | Line 6: | ||
<meta charset="utf-8" /> | <meta charset="utf-8" /> | ||
<title>InterLab</title> | <title>InterLab</title> | ||
| + | <script> | ||
| + | $(document).ready(function () { | ||
| + | $('#banner img').attr('src', 'https://static.igem.org/mediawiki/2018/f/f0/T--NAU-China--bannerdesign.jpg') | ||
| + | }); | ||
| + | </script> | ||
<style> | <style> | ||
| − | + | .top-title { | |
| − | . | + | color: #b6471d !important; |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
} | } | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
.main-content h1 { | .main-content h1 { | ||
font-family: 'Avenir Next Condensed',sans-serif; | font-family: 'Avenir Next Condensed',sans-serif; | ||
| − | color: # | + | color: #b6471d; |
| − | margin:32px 0 !important; | + | margin: 32px 0 !important; |
| − | margin-left:40px !important; | + | margin-left: 40px !important; |
| − | font-size:48px; | + | font-size: 48px; |
| − | font-weight:bold !important; | + | font-weight: bold !important; |
| + | line-height: 60px; | ||
} | } | ||
| − | + | figure div { | |
| − | + | text-align:center; | |
| − | + | font-size:0.9em; | |
| − | + | line-height:1.2em; | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | line-height:2em; | + | |
} | } | ||
</style> | </style> | ||
</head> | </head> | ||
<body> | <body> | ||
| − | < | + | <div class="topLine"> |
| − | <img src="https://static.igem.org/mediawiki/2018/ | + | <p class="top-title">PROJECT</p> |
| − | < | + | <p class="sec-title">Design</p> |
| − | </ | + | </div> |
| − | + | ||
| + | <a href="https://2018.igem.org/Team:NAU-CHINA/Overview"> | ||
| + | <img id="icon1" class="guide-icon" src="https://static.igem.org/mediawiki/2018/8/89/T--NAU-China--projectovervieworange.png" /> | ||
| + | </a> | ||
| + | <a href="https://2018.igem.org/Team:NAU-CHINA/InterLab"> | ||
| + | <img id="icon2" class="guide-icon" src="https://static.igem.org/mediawiki/2018/6/6a/T--NAU-China--interlaborange.png" /> | ||
| + | </a> | ||
| + | <a href="https://2018.igem.org/Team:NAU-CHINA/Protocols"> | ||
| + | <img id="icon3" class="guide-icon" src="https://static.igem.org/mediawiki/2018/3/3f/T--NAU-China--protocolorange.png" /> | ||
| + | </a> | ||
| + | <a href="https://2018.igem.org/Team:NAU-CHINA/Application_prospects"> | ||
| + | <img id="icon4" class="guide-icon" src="https://static.igem.org/mediawiki/2018/b/b0/T--NAU-China--iconapplicationorange.png" /> | ||
| + | </a> | ||
| + | |||
<div class="main-content"> | <div class="main-content"> | ||
<div class="textblock"> | <div class="textblock"> | ||
<h1>Introduction</h1> | <h1>Introduction</h1> | ||
| − | <p>MOSFET(metal-oxide-semiconductor field-effect transistor) is an essential component in both analog and digital circuits | + | <p>MOSFET (metal-oxide-semiconductor field-effect transistor) is an essential component in both analog and digital circuits, serving as analog switches or micro-processors. Inspired by this idea, we built genetic circuit "MOSFETs" in animal T cells, which is Monitoring and Operating System Founded on Engineered T cells. We hope our system can serve as a sensitive bioswitch to real-time monitor the extracellular concentration of a certain antigen, and determine whether to activate the expression of a downstream protein according to the preset threshold. As we expect, it should make no response to low concentration, but have quite high sensitivity near the threshold. In order to achieve our goal, we introduced synNotch, TetR-TetO, recombinase and Recombination Directionality Factors (RDF)in our system.Our MOSFET can be divided into 4 devices, namely, signal detection, signal processing, signal output and system reset.</p> |
| − | <img src="https://static.igem.org/mediawiki/2018/ | + | <img src="https://static.igem.org/mediawiki/2018/f/ff/T--NAU-China--xsmosfet.jpg" style="margin:0 auto;display:block;"> |
</div> | </div> | ||
| Line 91: | Line 62: | ||
<p> | <p> | ||
<b>SynNotch, an engineered transmembrane receptor, bridges intra- and extra-cellular information.</b> | <b>SynNotch, an engineered transmembrane receptor, bridges intra- and extra-cellular information.</b> | ||
| − | + | </p> | |
| − | + | <p>Synthetic Notch (SynNotch)[1]consists of three parts: the synthetic extracellular recognition domain (SynECD, e.g.scFv), the core transmembrane domain of wild Notch receptor[2], and the synthetic intracellular transcriptional domain (SynICyi5yD, e.g.SynTF). When the SynECD binds to its target surface antigen, induced cleavages take place on the core transmembrane domain of SynNotch, releasing the SynICD. The SynICD would be transported into the nucleus and activate the transcription of its corresponding gene.</p> | |
| − | + | <p><b>SynNotch is an ideal platform for customized antigen sensing behavior.</b></p> | |
| − | + | <p>SynNotch provides us an exciting platform because its SynECD and SynICD are both customizable. SynECD can be designed based on currently available scFvs for different tumors .SynICD will trigger customized output after SynECD recognition.</p> | |
| − | + | <figure> | |
| − | The triggering of SynNotch pathway | + | <img src="https://static.igem.org/mediawiki/2018/4/49/T--NAU-CHINA--xs100.jpg"> |
| − | translocating and promoting transcription. SynNotch is a customizable platform | + | <figcaption> |
| − | for cell sensing and response.</figcaption></figure> | + | <div>Fig.1 The logic of SynNotch </div> |
| − | + | The triggering of SynNotch pathway has 4 processes: antigen binding, cleavage, translocating and promoting transcription. SynNotch is a customizable platform for cell sensing and response. | |
| − | + | </figcaption> | |
| + | </figure> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/b/b7/T--NAU-CHINA--xssyn.jpg"> | ||
| + | <figcaption>Fig.2 Here, we replace the extracellular domain with anti-EGFP, which can recognize cell surface-expressed EGFP, and the incellular domain with TEV protease, which can cleave specific-designed TetR and activate the expression of recombinase.</figcaption> | ||
| + | </figure> | ||
</div> | </div> | ||
| − | |||
<div class="textblock"> | <div class="textblock"> | ||
<h1>Signal Processing</h1> | <h1>Signal Processing</h1> | ||
<div class="section"> | <div class="section"> | ||
| − | + | <p>We want to convert the extracellular analog signal into an intracellular digital signal, using Tet-Off/Tet-On device. This part contains tetO(Tetracycline operator), TEV protease (Tobacco Etch Virus nuclear-inclusion-a endopeptidase) and TetR(Tetracycline repressor protein).</p> | |
<p> | <p> | ||
| − | + | TetO is a operon which can be repressed by TetR.<br> | |
| + | TEV protease (Tobacco Etch Virus nuclear-inclusion-a endopeptidase), a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV) [4], is currently widely used in synthetic biology.<br> | ||
| + | TetR is a repressor from E. coli that blocks downstream expression when it binds to the TetO [3].In our project, we use specifically-designed TetR with TEV protease cleavage site which has been reformed by 2017 Oxford University iGEM project. | ||
</p> | </p> | ||
| − | <p> | + | <p>To our knowledge, the Tet control system is the perfect combination of prokaryotic and eukaryotic gene expression regulation systems.</p> |
| − | + | <figure> | |
| − | <figure><img src="https://static.igem.org/mediawiki/2018/ | + | <img src="https://static.igem.org/mediawiki/2018/7/7b/T--NAU-China--xs3.gif"> |
| − | <figcaption | + | <figcaption>Fig.3 TetR binds to the DNA operon and inhibits the production of export proteins. However, TetR has a cleavage site for Tobacco Etch Virus (TEV) protease. When it is cut by TEV, the suppression will be alleviated and the downstream gene could express.</figcaption> |
| − | + | </figure> | |
| − | + | <p>Initially,We thought that we can implement the filtering function using these devices. But, when we verificated system by the mathematical model, we found that there was a small amount of continuous tetO-downstream promoter leakage, when there was no extracellular signal stimulation. The leakage was 1% of the normal expression (fig.4), but this still need to be improved. To solve the problem, we introduced a recombinase into the system.</p> | |
| − | + | <figure> | |
| − | <figcaption | + | <img src="https://static.igem.org/mediawiki/2018/0/0d/T--NAU-China--xs4.jpg"> |
| − | + | <figcaption> | |
| − | + | <div>Fig.4 Expression of GFP in different TEV concentration levelswithout using recombinase</div> | |
| − | <figcaption | + | (As for each chart in this part, we choose the quantity of GFP as the vertical axis, and 0.01 hour as the unit of the horizontal axis. Different subplots correspond to different levels of intracellular protein concentration). It can be seen that under low signal levels, the effectors have inevitable leakage, which is not allowed by us, so the rec system have tobe used to ensure the rigor of the system.</figcaption> |
| − | + | </figure> | |
| − | + | <p>Recombinases , especially a subset called serine integrases and excisionases[5], are enzymes that can flip specific fragments of DNA. Recombinase can directionally catalyze sensitive DNA exchange reactions between targeted short (30–40 nucleotides) sequence sites that are specific to each recombinase[6]. They have been proved to be able to stably modify DNA sequences, which is the biological basis of our MOSFET construct. </p> | |
| − | <figcaption | + | <figure> |
| − | + | <img src="https://static.igem.org/mediawiki/2018/4/4a/T--NAU-China--xs5.gif"> | |
| − | + | <figcaption> | |
| − | + | <div>Fig. 5 Large serine integrases reliably and irreversibly flip unique fragments of DNA</div> | |
| − | + | DNA cleavage and re-ligation occur at the central crossover region at a pair of recombinase recognition sites (attB and attP), which allows the sequence to be flipped between recognition sites. After recombination, the original attB and attP sequences become reconstructed sequences attL and attR. The resulting attL and attR sequences cannot be recognized and bound by integrases alone, so the state after integration is stable.</figcaption> | |
| − | <h1> | + | </figure> |
| + | <h1>Signal output</h1> | ||
| + | <p>After mathematic model verification, it was found that the system achieved equal outputs with different thresholds and jump by using different promoters and recombinases. These features mean that it can function as quick-response switches. The Model team fed back to the Wet team, hoping to make more diverse switches by combining different promoters and recombinases.</p> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/c/c6/T--NAU-China--xs6.jpg"> | ||
| + | <figcaption> | ||
| + | <div>Fig. 6 We re-selected the tetO-downstream promoters</div> | ||
| + | UbC, EF1α, and miniCMV were selected and matched with different recombinases, including TP901, Bxb1 and PhiC31, by adjusting the parameters of each part and the strength of the promoter. Our whole system has formed nine threshold choices.</figcaption> | ||
| + | </figure> | ||
| + | <p>Theoretically, by selecting different promoters and recombinases, we can actually control whether the recombinase can successfully complete the reverse of its downstream sequence under a certain antigen concentration. In other words, we can preset the threshold antigen concentration according to our practical applications.</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="textblock"> | ||
| + | <h1>System reset</h1> | ||
<div class="section"> | <div class="section"> | ||
| − | + | <p>To realize a resettable and more accurate 0/1 switch, we introduce recombination directionality factor (RDF) to our system. When the external signal disappears or falls below the threshold, our MOSFET is able to reset to restore the initial state.</p> | |
| − | <p> | + | <p>In the signal processing part of our system, we have mentioned recombinases, which promote recombination between two different DNA sites, attP and attB, and in turn form recombinant attL and attR sites. The ‘reverse’ reaction requires another phage-encoded protein called RDF. RDF activates attL×attR recombination and inhibits attP×attB recombination. Recombinases can be fused to their corresponding RDFs to create single proteins that catalyse efficient attL×attR recombination in vivo and in vitro[7].</p> |
| − | + | ||
| − | < | + | <figure> |
| − | + | <img src="https://static.igem.org/mediawiki/2018/d/d3/T--NAU-CHINA--xsczm.jpg"> | |
| − | + | <figcaption><div>Fig. 7 RDF identifies the attL×attR site and flips it back to the original attP×attB state, which closes the device</div></figcaption> | |
| − | + | </figure> | |
| − | <figcaption | + | <p>Immediately, we started a new round of mathematical model verification. When the recombinase and RDF existed together, our system would be unstable. While the recombinase brought the system to the “0” state, the RDF would interfere with its behavior and make it into “1” state. (Fig.8). In order to enhance the stability of the system, we introduced the RDF-inhibitor part. RDF-inhibitor can interact with RDF and make it lose its flipping ability.In our scenario, RDF inhibitors can be siRNAs[8].</p> |
| − | + | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/1/1d/T--NAU-China--xs8.jpg"> | ||
| + | <figcaption> | ||
| + | <div>Fig. 8 Switching of downstream genes without RDF-inhibitor noise reduction</div> | ||
| + | X axis represents whether the downstream gene is active. We can see that switches of downstream gene are controlled by signal concentration. Only when concentration level is more than 5, the switch will be on. However, because the expression of RDF is not restricted, it will reset the downstream genes directly, which will close the switch. Then the gene will be open by the effect of rec again, which will cause periodical shock in the system. Because of the shock of effectors’ gene, the expression of GFP is discontinuous. When the system needs to be turned on, oscillations may occur, which may damage the DNA. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| − | <figure><img src="https://static.igem.org/mediawiki/2018/ | + | <figure> |
| − | <figcaption | + | <img src="https://static.igem.org/mediawiki/2018/5/50/T--NAU-CHINA--xs9.jpg"> |
| − | + | <figcaption>Fig.9 T-cells constitutively express tetR and the promoter is inhibited. When the cells sense the signal in the environment, TEV will bind to tetR, so the recombinase and the inhibitor could be expressed. </figcaption> | |
| − | + | </figure> | |
| − | + | <p>①When the TEV protease concentration reaches the threshold, the inhibition of the downstream gene by the TetR could be released. Then, the recombinase recognizes the attB and attP sites, reverses the sequence between attB and attP, and generates new attL and attR sites. The recombinase-RDF and RFP protein can be expressed after the reversal. But the recombinase-RDF loses function by binding with the inhibitor produced upstream. The RFP concentration continues to rise.</p> | |
| − | + | <p>②When the external signal weakens and the intracellular TEV protein concentration falls below the threshold, the TetR re-block the expression of recombinase and the inhibitor. Then recombinase-RDF recognizes the attL and attR sites and shuts down the expression of RFP protein. </p> | |
| − | + | <p>Based on the above design , we call it the resettable exact "0/1" switch</p> | |
| − | + | </div> | |
| − | + | </div> | |
<div class="textblock"> | <div class="textblock"> | ||
| − | <h1>Reference</h1> | + | <h1>Reference</h1> |
| − | <p>1 | + | <p>[1] Morsut, L. et al. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell164, 780–791 (2016).</p> |
| − | <p>2 | + | <p>[2] Bray, S. J. Notch signalling in context. Nat. Rev. Mol. Cell Biol.17, 722–735 (2016).</p> |
| − | <p>3 | + | <p>[3] Ramos, J. L. et al. The TetR Family of Transcriptional Repressors The TetR Family of Transcriptional Repressors. Microbiol. Mol. Biol. Rev.69, 326–356 (2005).</p> |
| − | + | <p>[4] Phan, J. et al. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem.277, 50564–50572 (2002).</p> | |
| − | <p>4 | + | <p>[5] Stark WM. 2014. The serine recombinases. Microbiol Spectrum 2(6):MDNA3-0046-2014.</p> |
| − | + | <p>[6] Nern, A., Pfeiffer, B. D., Svoboda, K. & Rubin, G. M. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl. Acad. Sci. 108, 14198–14203 (2011).</p> | |
| − | <p>5 | + | <p>[7] Olorunniji, F. J. et al. Control of serine integrase recombination directionality by fusion with the directionality factor. Nucleic Acids Res. 45, 8635–8645 (2017).</p> |
| − | + | <p>[8] Joshi, B. H. &Pachchigar, K. P. SiRNA: Novel therapeutics from functional genomics. Biotechnol. Genet. Eng. Rev. 30, 1–30 (2014).</p> | |
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
</body> | </body> | ||
</html> | </html> | ||
| + | {{NAU-CHINA/footer}} | ||
Latest revision as of 14:54, 3 November 2018

PROJECT
Design



Introduction
MOSFET (metal-oxide-semiconductor field-effect transistor) is an essential component in both analog and digital circuits, serving as analog switches or micro-processors. Inspired by this idea, we built genetic circuit "MOSFETs" in animal T cells, which is Monitoring and Operating System Founded on Engineered T cells. We hope our system can serve as a sensitive bioswitch to real-time monitor the extracellular concentration of a certain antigen, and determine whether to activate the expression of a downstream protein according to the preset threshold. As we expect, it should make no response to low concentration, but have quite high sensitivity near the threshold. In order to achieve our goal, we introduced synNotch, TetR-TetO, recombinase and Recombination Directionality Factors (RDF)in our system.Our MOSFET can be divided into 4 devices, namely, signal detection, signal processing, signal output and system reset.

Signal Detection
SynNotch, an engineered transmembrane receptor, bridges intra- and extra-cellular information.
Synthetic Notch (SynNotch)[1]consists of three parts: the synthetic extracellular recognition domain (SynECD, e.g.scFv), the core transmembrane domain of wild Notch receptor[2], and the synthetic intracellular transcriptional domain (SynICyi5yD, e.g.SynTF). When the SynECD binds to its target surface antigen, induced cleavages take place on the core transmembrane domain of SynNotch, releasing the SynICD. The SynICD would be transported into the nucleus and activate the transcription of its corresponding gene.
SynNotch is an ideal platform for customized antigen sensing behavior.
SynNotch provides us an exciting platform because its SynECD and SynICD are both customizable. SynECD can be designed based on currently available scFvs for different tumors .SynICD will trigger customized output after SynECD recognition.


Signal Processing
We want to convert the extracellular analog signal into an intracellular digital signal, using Tet-Off/Tet-On device. This part contains tetO(Tetracycline operator), TEV protease (Tobacco Etch Virus nuclear-inclusion-a endopeptidase) and TetR(Tetracycline repressor protein).
TetO is a operon which can be repressed by TetR.
TEV protease (Tobacco Etch Virus nuclear-inclusion-a endopeptidase), a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV) [4], is currently widely used in synthetic biology.
TetR is a repressor from E. coli that blocks downstream expression when it binds to the TetO [3].In our project, we use specifically-designed TetR with TEV protease cleavage site which has been reformed by 2017 Oxford University iGEM project.
To our knowledge, the Tet control system is the perfect combination of prokaryotic and eukaryotic gene expression regulation systems.

Initially,We thought that we can implement the filtering function using these devices. But, when we verificated system by the mathematical model, we found that there was a small amount of continuous tetO-downstream promoter leakage, when there was no extracellular signal stimulation. The leakage was 1% of the normal expression (fig.4), but this still need to be improved. To solve the problem, we introduced a recombinase into the system.

Recombinases , especially a subset called serine integrases and excisionases[5], are enzymes that can flip specific fragments of DNA. Recombinase can directionally catalyze sensitive DNA exchange reactions between targeted short (30–40 nucleotides) sequence sites that are specific to each recombinase[6]. They have been proved to be able to stably modify DNA sequences, which is the biological basis of our MOSFET construct.
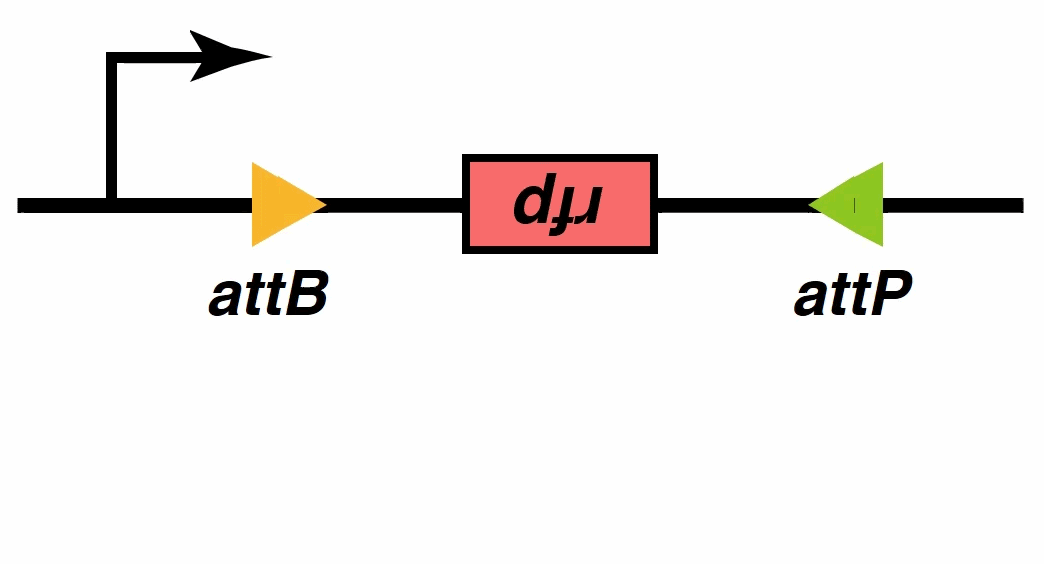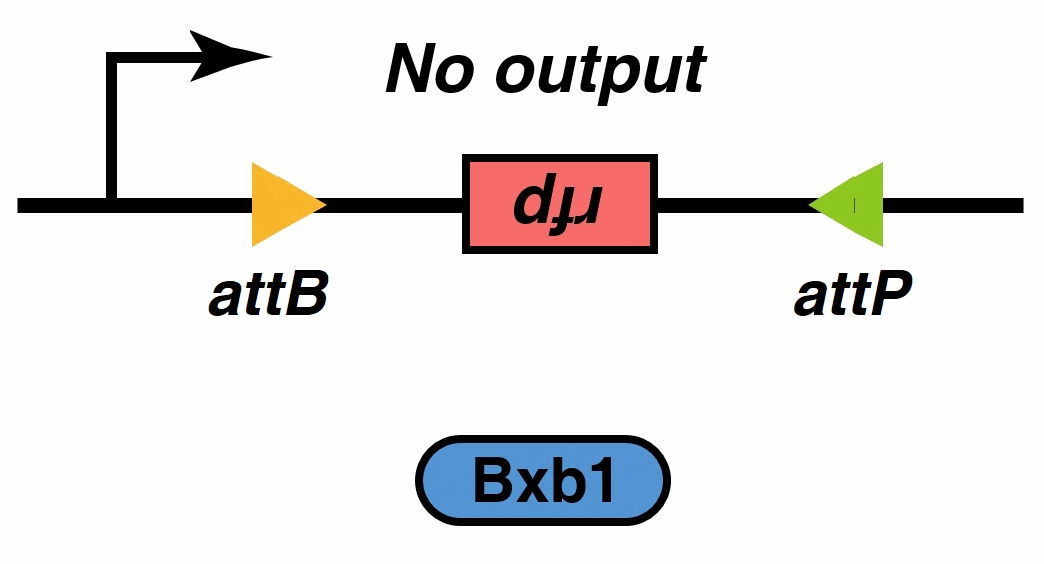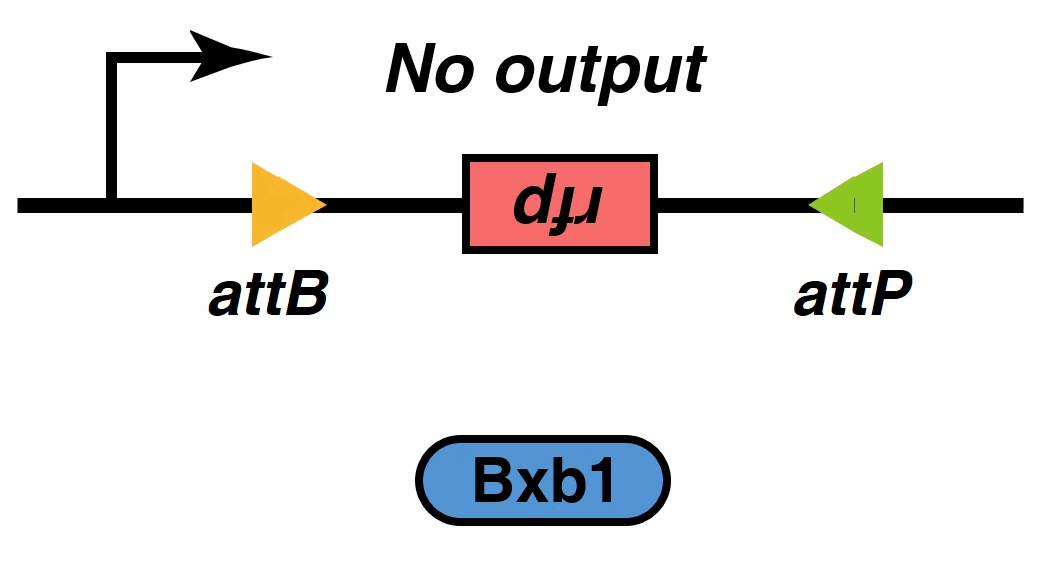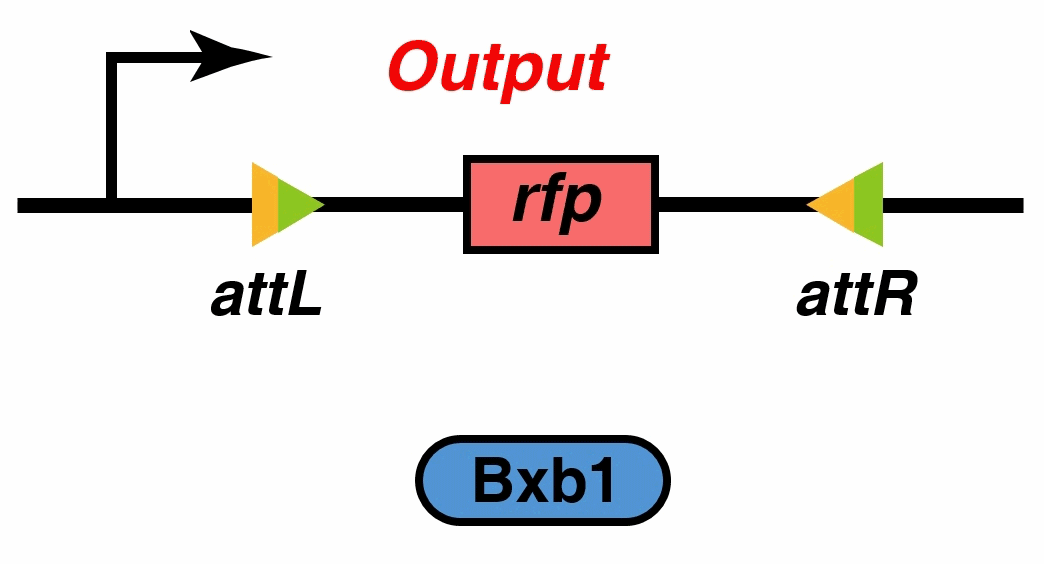
Signal output
After mathematic model verification, it was found that the system achieved equal outputs with different thresholds and jump by using different promoters and recombinases. These features mean that it can function as quick-response switches. The Model team fed back to the Wet team, hoping to make more diverse switches by combining different promoters and recombinases.

Theoretically, by selecting different promoters and recombinases, we can actually control whether the recombinase can successfully complete the reverse of its downstream sequence under a certain antigen concentration. In other words, we can preset the threshold antigen concentration according to our practical applications.
System reset
To realize a resettable and more accurate 0/1 switch, we introduce recombination directionality factor (RDF) to our system. When the external signal disappears or falls below the threshold, our MOSFET is able to reset to restore the initial state.
In the signal processing part of our system, we have mentioned recombinases, which promote recombination between two different DNA sites, attP and attB, and in turn form recombinant attL and attR sites. The ‘reverse’ reaction requires another phage-encoded protein called RDF. RDF activates attL×attR recombination and inhibits attP×attB recombination. Recombinases can be fused to their corresponding RDFs to create single proteins that catalyse efficient attL×attR recombination in vivo and in vitro[7].

Immediately, we started a new round of mathematical model verification. When the recombinase and RDF existed together, our system would be unstable. While the recombinase brought the system to the “0” state, the RDF would interfere with its behavior and make it into “1” state. (Fig.8). In order to enhance the stability of the system, we introduced the RDF-inhibitor part. RDF-inhibitor can interact with RDF and make it lose its flipping ability.In our scenario, RDF inhibitors can be siRNAs[8].


①When the TEV protease concentration reaches the threshold, the inhibition of the downstream gene by the TetR could be released. Then, the recombinase recognizes the attB and attP sites, reverses the sequence between attB and attP, and generates new attL and attR sites. The recombinase-RDF and RFP protein can be expressed after the reversal. But the recombinase-RDF loses function by binding with the inhibitor produced upstream. The RFP concentration continues to rise.
②When the external signal weakens and the intracellular TEV protein concentration falls below the threshold, the TetR re-block the expression of recombinase and the inhibitor. Then recombinase-RDF recognizes the attL and attR sites and shuts down the expression of RFP protein.
Based on the above design , we call it the resettable exact "0/1" switch
Reference
[1] Morsut, L. et al. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell164, 780–791 (2016).
[2] Bray, S. J. Notch signalling in context. Nat. Rev. Mol. Cell Biol.17, 722–735 (2016).
[3] Ramos, J. L. et al. The TetR Family of Transcriptional Repressors The TetR Family of Transcriptional Repressors. Microbiol. Mol. Biol. Rev.69, 326–356 (2005).
[4] Phan, J. et al. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem.277, 50564–50572 (2002).
[5] Stark WM. 2014. The serine recombinases. Microbiol Spectrum 2(6):MDNA3-0046-2014.
[6] Nern, A., Pfeiffer, B. D., Svoboda, K. & Rubin, G. M. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl. Acad. Sci. 108, 14198–14203 (2011).
[7] Olorunniji, F. J. et al. Control of serine integrase recombination directionality by fusion with the directionality factor. Nucleic Acids Res. 45, 8635–8645 (2017).
[8] Joshi, B. H. &Pachchigar, K. P. SiRNA: Novel therapeutics from functional genomics. Biotechnol. Genet. Eng. Rev. 30, 1–30 (2014).






