| (21 intermediate revisions by 4 users not shown) | |||
| Line 8: | Line 8: | ||
<meta content="width=device-width,initial-scale=1.0,maximum-scale=1.0,user-scalable=0;" name="viewport"> | <meta content="width=device-width,initial-scale=1.0,maximum-scale=1.0,user-scalable=0;" name="viewport"> | ||
<meta name="renderer" content="webkit"> | <meta name="renderer" content="webkit"> | ||
| − | <meta name="author" content="773715181 | + | <meta name="author" content="773715181 Jiaxiao Han"> |
<meta content="2018iGEM Team:Tianjin Kai System to control biological clock" name="description"> | <meta content="2018iGEM Team:Tianjin Kai System to control biological clock" name="description"> | ||
<meta content="Team:Tianjin,iGEM:Tianjin,iGEM,2018iGEM" name="keywords"> | <meta content="Team:Tianjin,iGEM:Tianjin,iGEM,2018iGEM" name="keywords"> | ||
| Line 15: | Line 15: | ||
<link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/bootstrap?action=raw&ctype=text/css"> | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/bootstrap?action=raw&ctype=text/css"> | ||
<link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/base?action=raw&ctype=text/css"> | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/base?action=raw&ctype=text/css"> | ||
| − | <link href="// | + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/font-awesome?action=raw&ctype=text/css"> |
<link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/text?action=raw&ctype=text/css"> | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/text?action=raw&ctype=text/css"> | ||
<script src="https://2018.igem.org/Team:Tianjin/js/bootstrap?action=raw&ctype=text/javascript"></script> | <script src="https://2018.igem.org/Team:Tianjin/js/bootstrap?action=raw&ctype=text/javascript"></script> | ||
| + | <link rel="stylesheet" type="text/css" href="https://2018.igem.org/Team:Tianjin/css/slides?action=raw&ctype=text/css"> | ||
</head> | </head> | ||
<!-- delete.css --> | <!-- delete.css --> | ||
| Line 28: | Line 29: | ||
.igem_2018_team_menu{display:none !important;} | .igem_2018_team_menu{display:none !important;} | ||
</style> | </style> | ||
| − | + | ||
<script> | <script> | ||
$('#top_title').remove();$("#globalWrapper").removeAttr("id");$("#content").removeAttr("id");$("#HQ_page").removeAttr("id");$('.mw-body').removeClass('mw-body');$('.igem_2018_team_mobile_bar').remove(); | $('#top_title').remove();$("#globalWrapper").removeAttr("id");$("#content").removeAttr("id");$("#HQ_page").removeAttr("id");$('.mw-body').removeClass('mw-body');$('.igem_2018_team_mobile_bar').remove(); | ||
| Line 62: | Line 63: | ||
</a> | </a> | ||
<ul class="dropdown-menu" style="left: -14px;"> | <ul class="dropdown-menu" style="left: -14px;"> | ||
| − | <li><a href="https://2018.igem.org/Team:Tianjin/Description"> | + | <li><a href="https://2018.igem.org/Team:Tianjin/Description">BACKGROUND</a></li> |
<li><a href="https://2018.igem.org/Team:Tianjin/Design">DESIGN</a></li> | <li><a href="https://2018.igem.org/Team:Tianjin/Design">DESIGN</a></li> | ||
<li><a href="https://2018.igem.org/Team:Tianjin/Experiments">EXPERIMENTS</a></li> | <li><a href="https://2018.igem.org/Team:Tianjin/Experiments">EXPERIMENTS</a></li> | ||
| Line 97: | Line 98: | ||
<li><a href="https://2018.igem.org/Team:Tianjin/Human_Practices">INTEGRATED</a></li> | <li><a href="https://2018.igem.org/Team:Tianjin/Human_Practices">INTEGRATED</a></li> | ||
<li><a href="https://2018.igem.org/Team:Tianjin/Collaborations">COLLABORATION</a></li> | <li><a href="https://2018.igem.org/Team:Tianjin/Collaborations">COLLABORATION</a></li> | ||
| − | <li><a href="https://2018.igem.org/Team:Tianjin/Public_Engagement"> | + | <li><a href="https://2018.igem.org/Team:Tianjin/Public_Engagement">PUBLIC ENGAGEMENT</a></li> |
</ul> | </ul> | ||
</li> | </li> | ||
| Line 111: | Line 112: | ||
</li> | </li> | ||
<li> | <li> | ||
| − | <a href="https://igem.org/ | + | <a href="https://2018.igem.org/Team:Tianjin/Judging"> |
| − | + | FOR JUDGES | |
</a> | </a> | ||
</li> | </li> | ||
| Line 120: | Line 121: | ||
</nav> | </nav> | ||
<script type="text/javascript" src="https://2018.igem.org/Team:Tianjin/js/navigation?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2018.igem.org/Team:Tianjin/js/navigation?action=raw&ctype=text/javascript"></script> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<div class="container "> | <div class="container "> | ||
<div class="row"> | <div class="row"> | ||
<div class="col-xs-12 titleBox"> | <div class="col-xs-12 titleBox"> | ||
<div class="title title-big"> | <div class="title title-big"> | ||
| − | <p> | + | <p>MEASUREMENT</p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 144: | Line 137: | ||
<p> | <p> | ||
In pursuit of a rigorously scientific result, we have adopted multiple measuring approaches throughout the whole project, among which two predominant methods described in extreme detail in wiki, would inspire relative researchers.<br><br> | In pursuit of a rigorously scientific result, we have adopted multiple measuring approaches throughout the whole project, among which two predominant methods described in extreme detail in wiki, would inspire relative researchers.<br><br> | ||
| − | As two sophisticated and renowned reporters in biotechnological domain, fluorescent protein and luciferase were opted to illustrate the effectiveness of our constructed oscillating system, detected by combined means and instrument. Mcherry and EYFP, two selected fluorescent reporters were measured by both fluorescence spectrophotometer and Leica DMi8 respectively. Leica DMi8 allows us to visually and intuitively see the expression of the fluorescent protein EYFP in Saccharomyces cerevisiae | + | As two sophisticated and renowned reporters in biotechnological domain, fluorescent protein and luciferase were opted to illustrate the effectiveness of our constructed oscillating system, detected by combined means and instrument. Mcherry and EYFP, two selected fluorescent reporters were measured by both fluorescence spectrophotometer and Leica DMi8 respectively. Leica DMi8 allows us to visually and intuitively see the expression of the fluorescent protein EYFP in <em>Saccharomyces cerevisiae</em> In terms of luciferase, in addition to commonly used firefly luciferase, a courageous attempt we made was to choose a novel luciferase NanoLuc and its live cell substrate in spite of no records of the usefulness in <em>Saccharomyces cerevisiae</em> before and obtained expected outcome successfully. |
</p> | </p> | ||
</div> | </div> | ||
| Line 163: | Line 156: | ||
<div class="col-xs-12 text"> | <div class="col-xs-12 text"> | ||
<p> | <p> | ||
| − | To achieve the best measurement of our | + | To achieve the best measurement of our oscillatory system, we chose two different kinds of reporter gene as our downstream reporters, fluorescent protein gene and luciferase gene, which are both widely used in the biotechnological labs. |
</p> | </p> | ||
</div> | </div> | ||
| Line 188: | Line 181: | ||
</table> | </table> | ||
</div> | </div> | ||
| − | <div class="col- | + | <div class="col-md-12 col-xs-12"> |
| + | <div class="wrap"> | ||
| + | <div class="photo-container"> | ||
| + | <div class="display-left" onclick="" style="right:0px"> | ||
| + | <a>❮</a> | ||
| + | </div> | ||
| + | <div class="Slides"> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/6/61/T--Tianjin--comic1.1.gif"> | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/3/30/T--Tianjin--comic1.2.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/8/89/T--Tianjin--comic1.3.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/c/cc/T--Tianjin--comic1.4.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/5/59/T--Tianjin--comic1.5.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/4/49/T--Tianjin--comic1.6.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/7/7a/T--Tianjin--comic1.7.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/8/86/T--Tianjin--comic1.8.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/2/2c/T--Tianjin--comic1.9.gif" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/e/e2/T--Tianjin--comic1.10.gif" > | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="display-right" onclick="" style="left:0px;"> | ||
| + | <a>❯</a> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="control"> | ||
| + | <div class="cr"><a><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-xs-12 picture"> | ||
<img src="https://static.igem.org/mediawiki/2018/4/40/T--Tianjin--measurement1.jpg"> | <img src="https://static.igem.org/mediawiki/2018/4/40/T--Tianjin--measurement1.jpg"> | ||
<p>Figure 1. Mechanism of fluorescent proteins</p> | <p>Figure 1. Mechanism of fluorescent proteins</p> | ||
| Line 201: | Line 250: | ||
<p> | <p> | ||
Luciferase is a popular choice as a reporter for gene expression because functional enzyme is created immediately upon translation and the assay is rapid, reliable and easy to perform.<sup><a href="#re1">[1,2]</a></sup> The fluorescent output of luciferase gene is due to the chemical reaction between luciferase and corresponding substrate, which can be easily detected by the luminoskan microplate reader.<br> | Luciferase is a popular choice as a reporter for gene expression because functional enzyme is created immediately upon translation and the assay is rapid, reliable and easy to perform.<sup><a href="#re1">[1,2]</a></sup> The fluorescent output of luciferase gene is due to the chemical reaction between luciferase and corresponding substrate, which can be easily detected by the luminoskan microplate reader.<br> | ||
| − | We used firefly luciferase(Fluc) gene purchased from Promega and | + | We used firefly luciferase(Fluc) gene purchased from Promega and NanoLuc luciferase(Nluc) gene submitted by Team Tuebingen 2015 as another two reporters for our KaiABC system. |
</p> | </p> | ||
</div> | </div> | ||
| Line 215: | Line 264: | ||
</tbody> | </tbody> | ||
</table> | </table> | ||
| + | </div> | ||
| + | <div class="col-md-12 col-xs-12"> | ||
| + | <div class="wrap"> | ||
| + | <div class="photo-container"> | ||
| + | <div class="display-left" onclick="" style="right:0px"> | ||
| + | <a>❮</a> | ||
| + | </div> | ||
| + | <div class="Slides"> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/6/68/T--Tianjin--comic2.1.jpg"> | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/3/37/T--Tianjin--comic2.2.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/5/5e/T--Tianjin--comic2.3.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/7/70/T--Tianjin--comic2.4.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/a/a7/T--Tianjin--comic2.5.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/4/49/T--Tianjin--comic2.6.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/0/04/T--Tianjin--comic2.7.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/0/08/T--Tianjin--comic2.8.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/9/94/T--Tianjin--comic2.9.jpg" > | ||
| + | </div> | ||
| + | <div class="mySlides"> | ||
| + | <img src="https://static.igem.org/mediawiki/2018/0/07/T--Tianjin--comic2.10.jpg" > | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="display-right" onclick="" style="left:0px;"> | ||
| + | <a>❯</a> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="control"> | ||
| + | <div class="cr"><a><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | <div class="cr"><a ><i class="fa fa-circle" aria-hidden="true"></i></a></div> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
<div class="col-xs-12 picture"> | <div class="col-xs-12 picture"> | ||
| Line 254: | Line 359: | ||
When reconstructed in <em>Escherichia coli</em>, KaiABC oscillator showed a periodicity of 24 hours<sup><a href="#re3">[3]</a></sup>. To best verify our system can function in <em>Saccharomyces cerevisiae</em> , we planned to measure our system using different reporter at <strong>3-hour intervals for 1 day, 2 days, 3 days respectively</strong><strong>.</strong><br> | When reconstructed in <em>Escherichia coli</em>, KaiABC oscillator showed a periodicity of 24 hours<sup><a href="#re3">[3]</a></sup>. To best verify our system can function in <em>Saccharomyces cerevisiae</em> , we planned to measure our system using different reporter at <strong>3-hour intervals for 1 day, 2 days, 3 days respectively</strong><strong>.</strong><br> | ||
As for the measurement of fluorescent proteins, we not only used the common detecting instrument, fourescence spectrophotometer, but also make an attempt to record the fluorescent variation via the cutting-edge Leica DMi8 inverted microscope.<br> | As for the measurement of fluorescent proteins, we not only used the common detecting instrument, fourescence spectrophotometer, but also make an attempt to record the fluorescent variation via the cutting-edge Leica DMi8 inverted microscope.<br> | ||
| − | Another thing worth mentioning is that, we bought all of the luciferase assay system from company Promega, among them there was a new kind of | + | Another thing worth mentioning is that, we bought all of the luciferase assay system from company Promega, among them there was a new kind of NanoLuc luciferase system----Nano-Glo<sup>®</sup> Live Cell Assay System, which allows experimenters to detect live cell luminescent signals without lytic process, suitable for the 3-day assay. Despite there is no reference indicating that this product, a commonly used assay system in mammalian cells, could be used in<em> </em><em>Saccharomyces cerevisiae</em>, we decided to take a try and have successfully proved its validity. Consequently, we chose Nluc solely to complete our long time luminescent detection . |
</p> | </p> | ||
</div> | </div> | ||
| Line 265: | Line 370: | ||
</thead> | </thead> | ||
<tbody> | <tbody> | ||
| − | <tr><tr><td rowspan="4" width="154"><p><strong>Experimental</strong></p><br><br><p><strong>Group</strong></p></td><td width="468"><p>pABaC +pCiRbS + | + | <tr><tr><td rowspan="4" width="154"><p><strong>Experimental</strong></p><br><br><p><strong>Group</strong></p></td><td width="468"><p>pABaC +pCiRbS +p1m</p></td></tr><tr><td width="468"><p>pABaC +pbCiRS +p1m</p></td></tr><tr><td width="468"><p>pABaC +pCiRbS+p1E</p></td></tr><tr><td width="468"><p>pABaC +pbCiRS +p1E</p></td></tr><tr><td rowspan="6" width="154"><p><strong>Negative</strong></p><br><br><p><strong>Control Group</strong></p></td><td width="468"><p>pABaC +p1m</p></td></tr><tr><td width="468"><p>pCiRbS +p1m</p></td></tr><tr><td width="468"><p>pbCiRS +p1m</p></td></tr><tr><td width="468"><p>pABaC +p1E</p></td></tr><tr><td width="468"><p>pbCiRS +p1E</p></td></tr><tr><td width="468"><p>pCiRbS p1E</p></td></tr> |
</tbody> | </tbody> | ||
</table> | </table> | ||
| Line 281: | Line 386: | ||
</thead> | </thead> | ||
<tbody> | <tbody> | ||
| − | <tr><td width="154"><p><strong>Experimental</strong></p><p><strong>Group</strong></p></td><td width="468"><p>pABaC +pCiRbS + | + | <tr><td width="154"><p><strong>Experimental</strong></p><p><strong>Group</strong></p></td><td width="468"><p>pABaC +pCiRbS +p1F</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pABaC +pbCiRS +p1F</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pABaC +pCiRbS +p2N</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pABaC +pbCiRS +p2N</p></td></tr><tr><td width="154"><p><strong>Negative</strong></p><p><strong>Control Group</strong></p></td><td width="468"><p>pABaC +p1F</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pbCiRS +p1F</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pCiRbS +p1F</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pABaC +p2N</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pbCiRS +p2N</p></td></tr><tr><td width="154"><p><strong> </strong></p></td><td width="468"><p>pCiRbS +p2N</p></td></tr> |
</tbody> | </tbody> | ||
</table> | </table> | ||
| Line 301: | Line 406: | ||
</thead> | </thead> | ||
<tbody> | <tbody> | ||
| − | <tr><td rowspan="4" width="154"><p><strong>Experimental</strong></p><p><strong>Group</strong></p></td><td width="468"><p>pABaC +pCiRbS + | + | <tr><td rowspan="4" width="154"><p><strong>Experimental</strong></p><p><strong>Group</strong></p></td><td width="468"><p>pABaC +pCiRbS +p1N</p></td></tr><tr><td width="468"><p>pABaC +pbCiRS +p1N</p></td></tr><tr><td width="468"><p>pABaC +pCiRbS +p2N</p></td></tr><tr><td width="468"><p>pABaC +pbCiRS 1+p2N</p></td></tr><tr><td rowspan="4" width="154"><p><strong>Control Group</strong></p></td><td width="468"><p>pABaC +p2N</p></td></tr><tr><td width="468"><p>pbCiRS +p2N</p></td></tr><tr><td width="468"><p>pCiRbS +p2N</p></td></tr><tr><td width="468"><p>TDH3P+NanoLuc</p></td></tr> |
</tbody> | </tbody> | ||
</table> | </table> | ||
| Line 317: | Line 422: | ||
</p> | </p> | ||
</div> | </div> | ||
| − | <div class="col-xs-12 picture"> | + | <div class="col-xs-12 picture" id="M1"> |
<img src="https://static.igem.org/mediawiki/2018/2/2e/T--Tianjin--EYFP123.jpg"> | <img src="https://static.igem.org/mediawiki/2018/2/2e/T--Tianjin--EYFP123.jpg"> | ||
| − | <p>Figure 6. pABaC +pCiRbS+ | + | <p>Figure 6. pABaC +pCiRbS+p1E photoed by normal microscope and Leica DMi8 </p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 328: | Line 433: | ||
</div> | </div> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 463: | Line 523: | ||
</div> | </div> | ||
<script type="text/javascript" src="https://2018.igem.org/Team:Tianjin/js/timeline?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2018.igem.org/Team:Tianjin/js/timeline?action=raw&ctype=text/javascript"></script> | ||
| + | <script src="https://2018.igem.org/Team:Tianjin/js/slides?action=raw&ctype=text/javascript"></script> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 13:47, 13 December 2018
<!DOCTYPE html>
MEASUREMENT
In pursuit of a rigorously scientific result, we have adopted multiple measuring approaches throughout the whole project, among which two predominant methods described in extreme detail in wiki, would inspire relative researchers.
As two sophisticated and renowned reporters in biotechnological domain, fluorescent protein and luciferase were opted to illustrate the effectiveness of our constructed oscillating system, detected by combined means and instrument. Mcherry and EYFP, two selected fluorescent reporters were measured by both fluorescence spectrophotometer and Leica DMi8 respectively. Leica DMi8 allows us to visually and intuitively see the expression of the fluorescent protein EYFP in Saccharomyces cerevisiae In terms of luciferase, in addition to commonly used firefly luciferase, a courageous attempt we made was to choose a novel luciferase NanoLuc and its live cell substrate in spite of no records of the usefulness in Saccharomyces cerevisiae before and obtained expected outcome successfully.
To achieve the best measurement of our oscillatory system, we chose two different kinds of reporter gene as our downstream reporters, fluorescent protein gene and luciferase gene, which are both widely used in the biotechnological labs.
Fluorescent protein
In search for the part kit provided by the iGEM headquarter, we found several available fluorescent proteins. Under the instruction of our fluorescent protein judging model, we finally chose mCherry and EYFP as our fluorescent protein reporter.
| Table 1. Chosen fluorescent proteins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
Protein | λem(nm) | λem(nm) | part serial number | site | |||||||
mCherry | 587 | 610 | Part:BBa_E2060 | 3-7L | |||||||
EYFP | 513 | 527 | Part:BBa_E2030 | 3-7J | |||||||
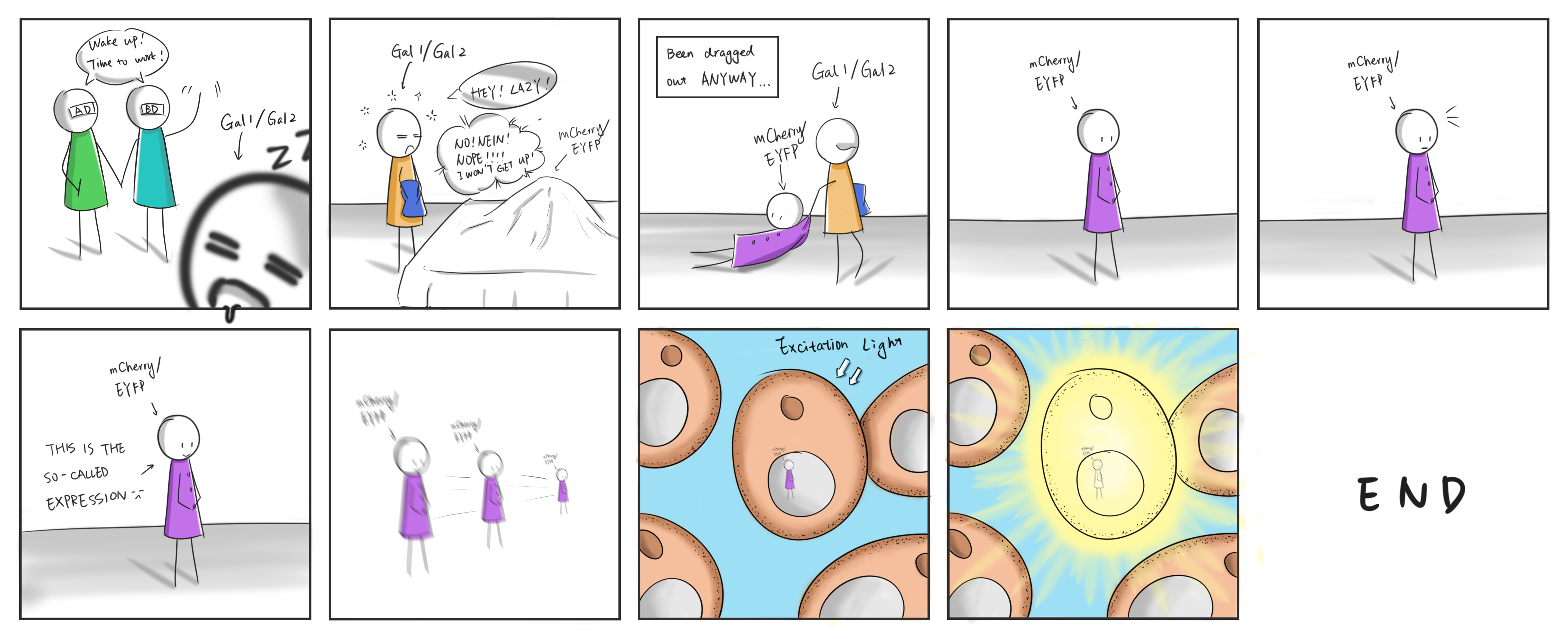
Figure 1. Mechanism of fluorescent proteins
Luciferase
Luciferase is a popular choice as a reporter for gene expression because functional enzyme is created immediately upon translation and the assay is rapid, reliable and easy to perform.[1,2] The fluorescent output of luciferase gene is due to the chemical reaction between luciferase and corresponding substrate, which can be easily detected by the luminoskan microplate reader.
We used firefly luciferase(Fluc) gene purchased from Promega and NanoLuc luciferase(Nluc) gene submitted by Team Tuebingen 2015 as another two reporters for our KaiABC system.
| Table 2. Chosen luciferase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
luciferase | λem(nm) | resource | Detail | ||||||||
Fluc | 560 | Bought from Promega | pGL4 Luciferase Reporter Vectors | ||||||||
Nluc | 460 | Part:BBa_K1680009 | Kit site:7-3N | ||||||||

Figure 2. Mechanism of luciferase

Figure 3.The bioluminescent reaction catalyzed by Fluc and Nluc
When reconstructed in Escherichia coli, KaiABC oscillator showed a periodicity of 24 hours[3]. To best verify our system can function in Saccharomyces cerevisiae , we planned to measure our system using different reporter at 3-hour intervals for 1 day, 2 days, 3 days respectively.
As for the measurement of fluorescent proteins, we not only used the common detecting instrument, fourescence spectrophotometer, but also make an attempt to record the fluorescent variation via the cutting-edge Leica DMi8 inverted microscope.
Another thing worth mentioning is that, we bought all of the luciferase assay system from company Promega, among them there was a new kind of NanoLuc luciferase system----Nano-Glo® Live Cell Assay System, which allows experimenters to detect live cell luminescent signals without lytic process, suitable for the 3-day assay. Despite there is no reference indicating that this product, a commonly used assay system in mammalian cells, could be used in Saccharomyces cerevisiae, we decided to take a try and have successfully proved its validity. Consequently, we chose Nluc solely to complete our long time luminescent detection .
| Table3.1 day’s measurement ----- Fluorescent protein detection | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
Experimental Group | pABaC +pCiRbS +p1m | ||||||||||
pABaC +pbCiRS +p1m | |||||||||||
pABaC +pCiRbS+p1E | |||||||||||
pABaC +pbCiRS +p1E | |||||||||||
Negative Control Group | pABaC +p1m | ||||||||||
pCiRbS +p1m | |||||||||||
pbCiRS +p1m | |||||||||||
pABaC +p1E | |||||||||||
pbCiRS +p1E | |||||||||||
pCiRbS p1E | |||||||||||

Figure 4. Procedures for measuring mCherry/EYFP via fluorescent spectrophotometer
| Table 4.2 days’ measurement ----- Fluc&Nluc detection in lytic cells | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
Experimental Group | pABaC +pCiRbS +p1F | ||||||||||
| pABaC +pbCiRS +p1F | ||||||||||
| pABaC +pCiRbS +p2N | ||||||||||
| pABaC +pbCiRS +p2N | ||||||||||
Negative Control Group | pABaC +p1F | ||||||||||
| pbCiRS +p1F | ||||||||||
| pCiRbS +p1F | ||||||||||
| pABaC +p2N | ||||||||||
| pbCiRS +p2N | ||||||||||
| pCiRbS +p2N | ||||||||||
Detailed methods are available in our Notebook.

Figure 5. Procedures for measuring luciferase via luminaskan microplate reader
| Table 5.3 days’ measurement ----- Nluc detection in live cells | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
Experimental Group | pABaC +pCiRbS +p1N | ||||||||||
pABaC +pbCiRS +p1N | |||||||||||
pABaC +pCiRbS +p2N | |||||||||||
pABaC +pbCiRS 1+p2N | |||||||||||
Control Group | pABaC +p2N | ||||||||||
pbCiRS +p2N | |||||||||||
pCiRbS +p2N | |||||||||||
TDH3P+NanoLuc | |||||||||||
EYFP measurement using Leica DMi8
Leica DMi8 is an excellent inverted microscope for life science research, which is utilized extensively in materials inspection and measurement. For live cell research, the DMi8 platform is a complete solution whether precisely follow the development of a single cell in a dish, screen through multiple assays, obtain single molecule resolution, or tease out behaviors of complex processes.
Combined with easy to use software, high resolution cameras, and brilliant LED illumination, these inverted research systems are flexible to see the expression of the fluorescent protein EYFP in Saccharomyces cerevisiae. Through high speed imaging, using the external fluorescent filter wheels, we obtained fast and accurate fluorescent images as following show. It tells that our bacteria that we constructed express EYFP successfully and continuously in oscillating system. Meanwhile, the fluorescence intensity detected was very bright, indicating that the expression of EYFP was at a high level.

Figure 6. pABaC +pCiRbS+p1E photoed by normal microscope and Leica DMi8




















