| Line 6: | Line 6: | ||
<img class="menuIcon" src="https://static.igem.org/mediawiki/2018/c/c0/T--Biomarvel--interlab.jpg"> | <img class="menuIcon" src="https://static.igem.org/mediawiki/2018/c/c0/T--Biomarvel--interlab.jpg"> | ||
<section id="mSection"> | <section id="mSection"> | ||
| − | <p>The InterLab study is organized by a measurement committee of iGEM to establish better measurement techniques in the synthetic biology field.We submit our measurement results that cope with the absorbance and fluorescence related | + | <p>The InterLab study is organized by a measurement committee of iGEM to establish better measurement techniques in the synthetic biology field. We submit our measurement results that cope with the absorbance and fluorescence related with <i>E. coli</i> transformed with a variety of test devices. We checked 8 plasmids, containing 2 controls and 6 test devices, and measurements were made in a plate reader with a 96 well microplate. We measured the absorbance and fluorescence of our samples using a Perkin Elmer 1420 multilabel counter.</p> |
</section> | </section> | ||
<section id="mSection"> | <section id="mSection"> | ||
| − | <h1>Calibration 1: OD <sub>600</sub> Reference | + | <h1>Calibration 1: OD <sub>600</sub> Reference Point - LUDOX Protocol</h1> |
| − | <p>We used LUDOX CL-X as a single point reference to obtain a conversion factor to transform absorbance (Abs <sub>600</sub>) data | + | <p>We used LUDOX CL-X as a single point reference to obtain a conversion factor to transform absorbance (Abs <sub>600</sub>) data from plate reader into acomparable OD <sub>600</sub> measurement as would be obtained in a spectrophotometer.</p> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/d/db/T--Biomarvel--inter1.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/d/db/T--Biomarvel--inter1.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 1. Measurement of Abs <sub>600</sub> and OD <sub>600</sub> for LUDOX CL-X and water</h4> |
</section> | </section> | ||
<section id="mSection"> | <section id="mSection"> | ||
<h1>Calibration 2: Particle Standard Curve - Microsphere Protocol</h1> | <h1>Calibration 2: Particle Standard Curve - Microsphere Protocol</h1> | ||
| − | <p>We prepared a dilution series of silica microspheres and | + | <p>We prepared a dilution series of silica microspheres and measured the Abs <sub>600</sub> in a plate reader. These microspheres are similar to cells in physical size and optical characteristics. This measurement was used to obtain a standardcurve of particle concentration, enabling to convert Abs <sub>600</sub> measurements to an estimated number of cells.</p> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/c/ce/T--Biomarvel--inter2.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/c/ce/T--Biomarvel--inter2.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 2. Calculated particle counts from measured Abs <sub>600</sub> and generation of a particle standard curve</h4> |
</section> | </section> | ||
<section id="mSection"> | <section id="mSection"> | ||
| − | <h1>Calibration 3: Fluorescence | + | <h1>Calibration 3: Fluorescence Standard Curve - Fluorescein Protocol</h1> |
| − | <p>We prepared a dilution series of fluorescein in four replicates and | + | <p>We prepared a dilution series of fluorescein in four replicates and measured the fluorescence in a 96 well plate in a plate reader. Measuring these in the plate readerenabled us togenerate astandard curve of fluorescence for fluorescein concentration.</p> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/c/ca/T--Biomarvel--inter3.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/c/ca/T--Biomarvel--inter3.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 3. Correspondence between fluorescence and fluorescein concentration</h4> |
</section> | </section> | ||
<section id="mSection"> | <section id="mSection"> | ||
| − | <h1>Cell | + | <h1>Cell Measurement</h1> |
| − | <p>We had two plates to read at the end of procedure and one plate for each time point: 0 and 6 hours. On each plate,we | + | <p>We had two plates to read at the end of procedure and one plate for each time point: 0 and 6 hours. On each plate, we read both fluorescence and absorbance. We used the generated particle standard and fluorescent curves to determine fluorescence per OD and fluorescence per particle values.</p> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/e/ef/T--Biomarvel--inter4.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/e/ef/T--Biomarvel--inter4.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 4. Measurement of raw Abs <sub>600</sub> and OD <sub>600</sub></h4> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/8/8c/T--Biomarvel--inter5.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/8/8c/T--Biomarvel--inter5.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 5. Measurement of fluorescence per OD</h4> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/e/e0/T--Biomarvel--inter6.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/e/e0/T--Biomarvel--inter6.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 6. Measurement of fluorescence per particle</h4> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/d/de/T--Biomarvel--inter7.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/d/de/T--Biomarvel--inter7.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 7. The number of colonies and CFU/mL value in a starting sample of OD <sub>600</sub> = 0.1</h4> |
</section> | </section> | ||
<section id="mSection"> | <section id="mSection"> | ||
| − | <h1>Colony Forming Units per 0.1 OD<sub>600</sub><i>E. coli</i> cultures</h1> | + | <h1>Colony Forming Units per 0.1 OD <sub>600</sub> <i>E. coli</i> cultures</h1> |
| − | <p>For the CFU protocol, | + | <p>For the CFU protocol, we calculated colonies for our two Positive Control (BBa_I20270) cultures and our two negative Control (BBa_R0040) cultures. Our overnight cultures were diluted to yield a 0.1 OD <sub>600</sub> using calculations described in iGEM 2018 InterLab Study Protocol. After a serial dilution for each overnight culture, we plated the 8*10<sup>-3</sup>, 8*10<sup>-4</sup>, and 8*10<sup>-5</sup> dilutions on LB agar with triplication. With the assumption that one bacterial cell leads to one colony, we enumerated the number of colonies on each plate and multiplied the colony count by the final dilution factor to assess a CFU/mL value for a starting sample of OD <sub>600</sub>.</p> |
<img class="myimg" src="https://static.igem.org/mediawiki/2018/e/e2/T--Biomarvel--inter8.png"> | <img class="myimg" src="https://static.igem.org/mediawiki/2018/e/e2/T--Biomarvel--inter8.png"> | ||
| − | <h4 style="text-align: center;"> | + | <h4 style="text-align: center;">Figure 8. Plated DH5α cells of serial dilutions for starting samples</h4> |
</section> | </section> | ||
</div> | </div> | ||
Revision as of 12:10, 13 October 2018

The InterLab study is organized by a measurement committee of iGEM to establish better measurement techniques in the synthetic biology field. We submit our measurement results that cope with the absorbance and fluorescence related with E. coli transformed with a variety of test devices. We checked 8 plasmids, containing 2 controls and 6 test devices, and measurements were made in a plate reader with a 96 well microplate. We measured the absorbance and fluorescence of our samples using a Perkin Elmer 1420 multilabel counter.
Calibration 1: OD 600 Reference Point - LUDOX Protocol
We used LUDOX CL-X as a single point reference to obtain a conversion factor to transform absorbance (Abs 600) data from plate reader into acomparable OD 600 measurement as would be obtained in a spectrophotometer.

Figure 1. Measurement of Abs 600 and OD 600 for LUDOX CL-X and water
Calibration 2: Particle Standard Curve - Microsphere Protocol
We prepared a dilution series of silica microspheres and measured the Abs 600 in a plate reader. These microspheres are similar to cells in physical size and optical characteristics. This measurement was used to obtain a standardcurve of particle concentration, enabling to convert Abs 600 measurements to an estimated number of cells.
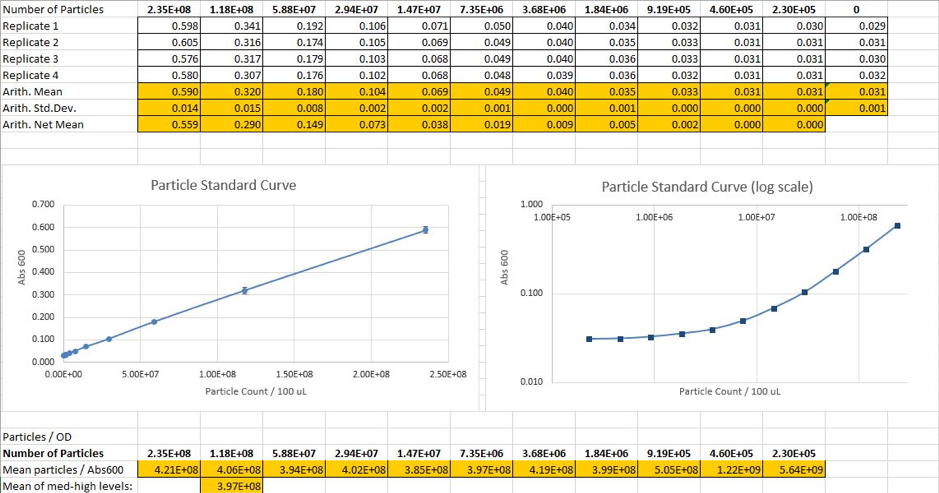
Figure 2. Calculated particle counts from measured Abs 600 and generation of a particle standard curve
Calibration 3: Fluorescence Standard Curve - Fluorescein Protocol
We prepared a dilution series of fluorescein in four replicates and measured the fluorescence in a 96 well plate in a plate reader. Measuring these in the plate readerenabled us togenerate astandard curve of fluorescence for fluorescein concentration.

Figure 3. Correspondence between fluorescence and fluorescein concentration
Cell Measurement
We had two plates to read at the end of procedure and one plate for each time point: 0 and 6 hours. On each plate, we read both fluorescence and absorbance. We used the generated particle standard and fluorescent curves to determine fluorescence per OD and fluorescence per particle values.

Figure 4. Measurement of raw Abs 600 and OD 600
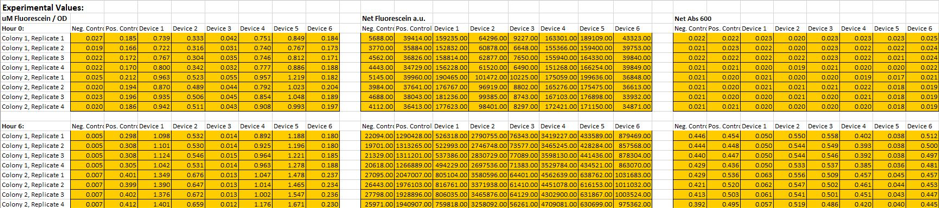
Figure 5. Measurement of fluorescence per OD

Figure 6. Measurement of fluorescence per particle

Figure 7. The number of colonies and CFU/mL value in a starting sample of OD 600 = 0.1
Colony Forming Units per 0.1 OD 600 E. coli cultures
For the CFU protocol, we calculated colonies for our two Positive Control (BBa_I20270) cultures and our two negative Control (BBa_R0040) cultures. Our overnight cultures were diluted to yield a 0.1 OD 600 using calculations described in iGEM 2018 InterLab Study Protocol. After a serial dilution for each overnight culture, we plated the 8*10-3, 8*10-4, and 8*10-5 dilutions on LB agar with triplication. With the assumption that one bacterial cell leads to one colony, we enumerated the number of colonies on each plate and multiplied the colony count by the final dilution factor to assess a CFU/mL value for a starting sample of OD 600.








