| Line 12: | Line 12: | ||
<meta charset="utf-8"> | <meta charset="utf-8"> | ||
<!--[if lt IE 9]> | <!--[if lt IE 9]> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<meta name="viewport" content="width=device-width, initial-scale=1, maximum-scale=1, user-scalable=0"> | <meta name="viewport" content="width=device-width, initial-scale=1, maximum-scale=1, user-scalable=0"> | ||
<!-- css bootstrap --> | <!-- css bootstrap --> | ||
Latest revision as of 01:56, 18 October 2018

Demonstration
- ✓ Geraniol production by E. coli
- ✓ Conversion of geraniol to nepetalactol by S. cerevisiae
- ✓ Creating a stable microbial consortium of E. coli and S. cerevisiae

Figure. 1 An overview of mCATNIP: distributing the biosynthesis pathway of nepetalactol between E. coli and S. cerevisiae
Geraniol Production by E. coli

Figure. 2 Geraniol production analysis by gas chromatography. (A) GPPS and GES are arranged in operon regulated by pTac, placed on a high copy vector pUC20. (B) The MVA pathway is split into two clusters and placed on a low copy vector with the upper cluster containing three genes and the downstream containing four. (C) A combined plasmid with GPPS&GES operon and MVA pathway with a low copy p15A origin. (D) Gas chromatography for geraniol produced by E. coli expressing pMVA only (middle trace) or pMVA-GPPS-GES (bottom trace) whose peaks coincide with a geraniol standard (top trace). (E) Geraniol yield is noticeable with only the heterologous MVA pathway. Upon introduction of GPPS and GES, the yield doubled relative to the negative control pMVA only.
We have proposed two different strategies for engineering a geraniol-producing E. coli strain: over-expressing GPPS and GES, which relies on native E. coli MEP pathway to supply IPP and DMAPP; and introducing a heterologous yeast MVA pathway along with GPPS and GES. For realizing these designs, we constructed (Figure. 1 A) pUC20-pTac-GPPS-GES and let Genscript constructed (Figure. 1 C) pMVA-GPPS-GES, and (Figure. 1 B) negative control pMVA for us. After transforming them into E. coli, we carried out a 24h shake-flask fermentation upon 25μM IPTG induction at a point when the OD600 equaled 1. Then, an n-hexane phase is used for harvesting geraniol from the culture which was analyzed by gas-chromatography.
The E. coli strain containing pUC20-pTac-GPPS-GES didn’t produce any detectable geraniol titer. Expression of pMVA-GPPS-GES yielded 11.4mg/L geraniol whereas expression of pMVA alone also produced 5.65mg/L geraniol. From this phenomenon we inferred that provision of the precursors IPP and DMAPP by heterologous pathway was necessary for achieving high geraniol yield.
However, we have not yet explored whether this expression level of GPPS and GES was the optimum at this condition. If the expression of GPPS and GES were tuned to suit the flux of precursors, we might see even greater titer of geraniol produced. Nonetheless, although at this moment we aimed at geraniol production by E. coli alone, the E. coli would eventually be co-cultured with S. cerevisiae for the production of nepetalactol. Then, since the change in culturing conditions has agitated the internal environment of the host, the pre-set ratio might be lost. Transcription-activator-like effector stabilized promoters (TALE sp) provided a solution to this problem through a regulatory mechanism employing the incoherent feedforward loop (iFFL).
We decided to utilize TALE sp to uncover the most suitable expression level of GPPS and GES. But the current number of TALE sp available was too small for us to make sensible decisions, which was the reason why we expanded the TALE sp library.

Figure. 3 Fluorescence characterization of the transcription-activator-like effector stabilized promoter (TALE sp) family. (A) Sequences of core promoter variants. TALE1 and TALE2 mean the TALE used this the promoter for negative regulation. Colored sequences refer to TALE binding sites. Names of the TALE sps harboring each core promoters are placed on the left of respective sequences. (B) Illustration for genetic parts design used in characterization. TALE1sp and TALE2sp are representatives of core promoters shown in (A). Identical characterization elements which include a RiboJ, an RBS, a sfGFP, and a terminator are placed downstream of TALE sps as well as J23 constitutive promoters for reference. (C) Performance of J23119, J23101, J23105 constitutive promoters on three plasmids with different copy numbers. The x-axis shows the reported copy number of plasmids used. (D) Performance of TALE1 stabilized promoters on the same set of plasmids used for characterizing constitutive promoters. Identities of the promoters are shown as legends to the right. In comparison to the sloped graph of J23 promoters, TALE1sps generate trend line that is more horizontal. (E) Performance of TALE2 stabilized promoters on the same set of plasmids used for characterizing constitutive promoters. Identities of the promoters are shown as legends to the right.
The expansion of the library was based on introducing a number of new core promoters — the TALE-repressible promoters (Figure. 2 A), and assembling them with the genetic circuit encoding for TALE (Figure. 2 B). After adding six more TALE sp, the library then had twelve members, whose performance was characterization by fluorescence given out by the sfGFP they regulated. Besides the TALE sp, we also characterized three constitutive promoters from the J23 consensus promoter family. (Figure. 2 C) Each promoter was placed on three backbones: the high copy pSB1C3, the medium copy pR6K, and the low copy pSC101, so that we could assess the stabilization ability. From the results measured by flow cytometry, we could see that the influence of copy number on fluorescence level has sharply decreased, signifying valid stabilization by TALE sp.
From the library, we chose three TALE sp to geraniol synthesis which were TALE2sp6, TALE2 sp1 and TALE1 sp1 in ascending promoter strength. They along with a pTac promoter were used for driving gene expression of GPPS and GES. Similar to the fluorescence characterization, the constructs were placed on three different vectors of high, medium and low copy. As MVA pathway was a requisite for geraniol production from our previous experience, we transferred the plasmids into the E. coli strain already harboring pMVA that we have created. Because pMVA contained chloramphenicol resistance which was the same as pSB1C3, pUC20 was chosen as the high copy vector for geraniol production assay.

Figure. 4 Geraniol production assay of TALE sp relative to pTac IPTG-inducible promoter. (A) Twelve combinations of promoters and plasmid backbones used to express a geraniol synthesis operon for characterization. pSC101 is the low copy vector, pR6K is the medium copy vector and pUC20 is the high copy vector. The combinations are transformed and tested in E. coli already harboring pMVA plasmids. (B) Geraniol yield achieved under the regulation of pTac, TALE1 sp1, TALE2 sp1, and TALE2 sp6 across different copy numbers is shown. The vertical error bars represent standard deviations of geraniol yield measured, and the horizontal error bars signify the reported range of copy number of plasmid backbones used. The portion of the graph belong to TALE2sp6 is not shown as there is no detectable geraniol production.
Table. 1 Geraniol production by different promoters and vector combinations
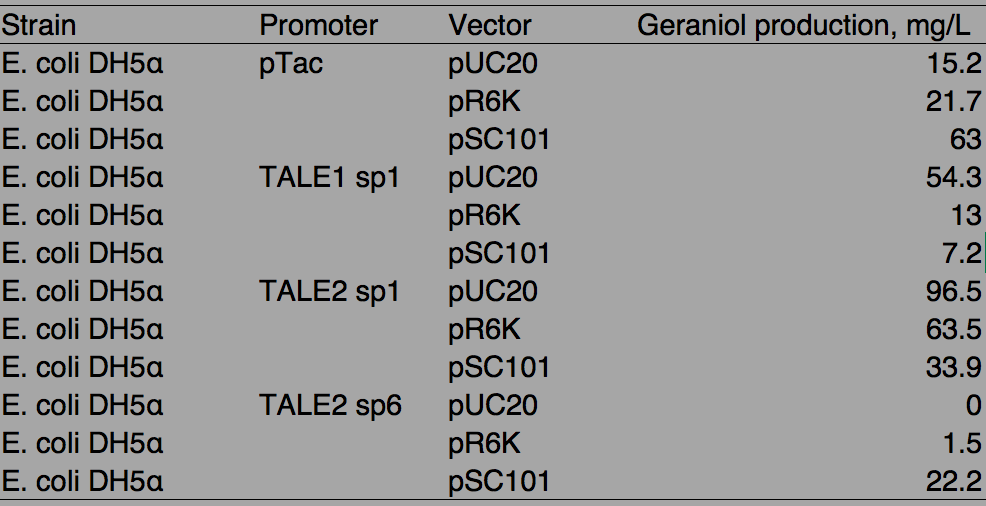
With pTac promoters, geraniol yield increased with the copy number of the vector, showing positively correlated relation. However, when TALE stabilized promoter (TALE sp) was used, higher the copy number of the vector, lower the production of geraniol, being the opposite of pTac. And the stronger TALE sp promoter, TALE1sp1 gave generally reduced yield compared to its weaker counterpart TALE2sp1. TALE2 sp6 whose strength measure with fluorescence is about half of the strength of TALE2 sp1 produced the lowest titre among the four promoters tested. Interestingly, the graphs of the TALE stabilised promoters appeared to be seemingly parallel to each other.
We surmised that the yield of geraniol was affected by two factors: the expression level of enzymes and the cellular burden. As for enzyme expression, there exists an optimal gene expression level that produces just enough enzyme to metabolize all the substrate. The yield would be the greatest at this level. But if lower than this level, production would increase with enzyme expression since there is a surplus of substrates. And if the enzyme expression is higher, the more enzymes now becomes a cellular burden as it no longer contributes to more product.

Figure. 5 Gas chromatography analysis for geraniol produced regulated by pTac and TALE2sp1 compared with the peak of geraniol standard. (A) Geraniol production accomplished by pTac regulation across low (pSC101), medium (pR6K) and high (pUC20) copy number. The area of the peak which appears at around 4.41s concurring with that of the geraniol standard increases along the copy number. (B) Geraniol yield assay using TALE2sp1 with the same set of copy number. The largest peak of geraniol emerges on the low copy vector pSC101. The peak shrinks in size on higher copy.
In the case of pTac, it fits into the scenario when the gene expression is lower than the optimum: higher copy produced more enzymes to catalyze geraniol synthesis reaction. As for TALE stabilized promoters, the expression level of enzymes would remain unchanged regardless of copy number due to the stabilization nature of TALE sp (Go to Design). So in low copy vector where the cellular burden isn’t significant, the product yield is only related to the strength of the promoter, explaining why TALE2 sp1 has better performance than both TALE1 sp1 and TALE2 sp6 as it’s closer to the optimum. But when the copy number increases, the expression level is unaffected while the cellular burden rises sharply because much more TALE would be made to stabilize expression, leaving less energy available for geraniol synthesis. And it in turns answers why a negative trend of production is shown when regulated by TALE sp.
It was reported that TALE stabilized promoters could maintain the level of gene expression without the need for “re-tuning” upon genomic insertion, so we used pOSIP to integrate TALE1sp1, the core promoter of TALE1sp1, TALE2sp1, and the core promoter of TALE2sp1 with the sfGFP into the attB locus in the genome individually and characterized the fluorescence again. The result shows that after genomic insertion, TALE stabilized promoters maintain a higher expression level in comparison to the core promoters which are constitutive promoters. Also, we inserted TALE2sp1-GPPS-GES into the genome to investigate if the trend of geraniol yield increasing on lower copy stayed consistent. In the genome where theoretic copy number is one, geraniol yield rose slightly from the previous level on pSC101, reaching 94.5 mg/L.

Figure. 6 Influence of genomic insertion upon the functionality of TALE stabilized promoters. (A) Fluorescence characterization of TALE1 sp1, TALE2 sp1 and their core promoters inserted to the same locus in the genome. TALE1 sp1 and TALE2 sp1 are shown in brown whereas their core promoters are in green. (B) The change in geraniol yield across different copy number when using TALE2sp1. From left to right, the first point presents the geraniol obtained then the operon is inserted to the genome. The second, the third and the fourth are values of geraniol titer on pSC101, pR6K, and pUC20 respectively.
Conversion of geraniol to nepetalactol by S. cerevisiae
From literature we have comprehended that fact that the conversion was unlikely unless promiscuous endogenous enzymes were knocked out, so we began expression vector construction and gene deletion at the same time.
We constructed pCRCT-OYE2, pCRCT-OYE3, and pCRCT-ADH7 which were three CRISPR-Cas9 plasmids targeting OYE2, OYE3, and ADH7 individually. Since an 8bp deletion is not visible using gel electrophoresis, and must be confirmed by sequencing, we made a small alteration in the design of homologous recombination disruption donors for knocking out OYE2 to make a 100bp deletion. In this way, the result of gene disruption would be easily determined using gel electrophoresis. We achieved OYE2 deletion through this method, but failed when applying it to knock out OYE3 and ADH7, so the former approach of removing 8bp was used instead. After few cycles of transforming pCRCT, confirming the deletion, and discarding pCRCT, we obtained an S. cerevisiae strain with all three genes knocked out.

Figure. 7 Sequencing results confirmed a successful disruption of oye2, oye3, and adh7. 100bp of OYE2 coding sequence is removed. As for OYE3 and ADH7, an 8bp deletion is generated.
Conversion of geraniol to nepetalactol by S. cerevisiae
As for the expression vector, at the beginning we used E. coli as the cloning host for constructing the vector, but to our surprise, no matter what assembly method we used — Gibson or biobrick assembly, the E. coli colony grown always missed the G8H component. After careful debug, we found that pTDH3 contains wild-type E. coli promoters which have triggered the expression of G8H in E. coli, resulting in cytotoxicity which forced E. coli to cleave G8H. So later we changed out a method by transforming the ligated DNA directly into yeast and using colony PCR to determine if the construction was successful.

Figure. 8 The results of the conversion of geraniol to nepetalactol. (A) S. cerevisiae expression vector with three genetic circuits encoding for G8H, GOR, and ISY on a 2 micron backbone. (B) Colony PCR gel electrophoresis image. Bands of correct length are seen meaning the construct was successful. (C) Gas chromatography results of geraniol consumption and conversion to nepetalactol. (1) is geraniol. (2),(3),and (4) are isomers of nepetalactol. (5) is an unknown products of metabolism. (D) The standard curve plotting to quantify nepetalactol yield
When we have eventually acquired the expression vector, the gene oye2 was knocked out. Therefore we started the characterization of using engineered yeast to convert geraniol to nepetalatcol. Having transferred the vector into both BY4741 and △oye2 BY4741, we shared the yeast at 30℃ and fed them with 40μl 200mM geraniol every two hours for 24h, then used gas chromatography for biocatalytic activity analysis. From the GC result, BY4741 has not produced nepetalactol, but we verified that geraniol was consumed by geraniol and nepetalactol was produced by △oye2 BY4741. However, an unknown metabolite was also generated and in a larger quantity than nepetalactol. Our assumption was that this peak was a shunt product reported. And if oye3 and adh7 were deleted in addition to oye2, it would be very likely that the nepetalactol yield increased and the shunt product decreased. At this moment, the nepetalactol yield we have obtained is 1.267mg/L.
Creating a stable microbial consortium of E. coli and S. cerevisiae
Co-culturing E. coli and S. cerevisiae has been a challenging task due to their different doubling time, optimal growth conditions, and ethanol excretion of S. cerevisiae inhibiting E. coli growth. Previous studies reveal a mechanism that prevents the competition between E. coli and S. cerevisiae to maintain stability in a co-culture system: using xylose as the carbon source. When xylose is served as the carbon source, only E. coli can directly utilize it but S. cerevisiae can not. Hence the only way for S. cerevisiae to survive is to use acetate, the waste product of the xylose metabolism in E. coli, as its carbon source. In addition, acetate inhibits the growth of E. coli so in this way E. coli and S. cerevisiae are mutually dependent on each other, which is known as commensalism.

Figure. 9 Ratio of E. coli and S. cerevisiae in the co-culture system given different carbon sources in 24h measured by flow cytometry. Flow cytometry sorts E. coli and S. cerevisiae into two piles. The lower region represents E. coli and the upper region represents S. cerevisiae. (A) When xylose is used as the carbon source, the percentages of S. cerevisiae at different time are: 2h, 34.3%; 4h, 25.5%; 6h, 27.9%; 8h, 20.83%; 10h, 8.14%; 12h, 2.66%; 14h, 2.59%; 16h, 2.43%; 18h, 1.71%; 20h, 1.80%; 22h, 1.97%; 24h, 1.88%. (B) The percentages of S. cerevisiae in 24h when fed glucose are: 2h, 23.47%; 4h, 38.91%; 6h, 54.90%; 8h, 73.85%; 10h, 77.02%; 12h, 75.03%; 14h, 75.59%; 16h, 74.68%; 18h, 72.59%; 20h, 72.34%; 22h, 66.92%; 24h, 63.59%.
We have examined the use of two carbon sources, glucose and xylose, to create a stable co-culture system of E. coli and S. cerevisiae.
In the first trial when glucose is fed, S. cerevisiae is the dominant party in the system during the first 24h. It started with a percentage of 23.47%, increased to 77.02% in the tenth hour, and dropped to 63.59% later at the twenty-fourth hour. However, we cannot conclude that the microbial consortium has reached a steady state as the percentage of S. cerevisiae kept fluctuating, and has shown a decreasing trend at the end. Longer monitoring might be needed in order to draw a valid conclusion.
And upon the use of xylose, E. coli became the advantageous species. The percentage of S. cerevisiae decreased right after the inoculation, but the proportion of it has remained relatively constant around 1.84% since 12h, from which we could know that the microbial consortium reached stability, proving the validity of xylose upon maintaining the balance in an E. coli and S. cerevisiae consortium.
In the future, if the proportion of S. cerevisiae is to be increased, we can enhance acetate supply through inactivation of the gene atpFH in E. coli.















