We intend to engineer E. coli DH5a to be the A. hydrophila killer. Our project can be fulfilled by three devices: 1). a sensor which can detect the presence of A. hydrophila in fish excrement and express green fluorescence protein. After farmers use fluorescence detector (only cost 60 RMB) to check the feces sample, they can deduce the amount of A. hydrophila in a fish body and take actions to avoid the outbreak of infection. 2). a primary helper killer which can express quorum quenching enzymes and reduce the virulence of A. hydrophila. 3). a secondary killer which expresses and releases antimicrobial peptides that can kill A. hydrophila directly. Thus, there are three kinds of killers with each device. The killers are put together into capsules and are mixed with fish fodder. After fish eat the capsules, the engineered E. coli can be released in its intestine, which is the organ A. hydrophila firstly invades and multiplies during infection. The vector we chose was Repressor Generator (RPG), which has p15A as the replication origin and ampicillin resistence. The copy number of the vector is among 20 per cell. We chose Prhl (LR) as the promoter to initiate the expression of GFP because it can be induced by C4-HSL, which is the most abundant quorum sensing molecule in A. hydrophila culture. rhlR (LVA-tagged) is the repressor & activator of the promoter, which represses the expression of GFP at low C4-HSL concentration but activates the expression at high C4-HSL concentration. The RBS of rhlR (LVA-tagged) and GFP is BBa_B0034, which is the strongest promoter documented in the part registry. However, since the downstream gene can affect the efficiency of RBS, when we used RBS calculator in salislab.net to predict the strength of these two RBS, we found the relative strength of BBa_B0034 on rhlR site and GFP site is 1:5. The experiment and model results of this device were not satisfying enough as well (see our demo). By using RBS calculator, we increased the strength of RBS of rhlR (LVA-tagged) to 5 times of the original one and measured the time-averaged GFP production rate of it. But since the device can only give 12.2% more fluorescence compared with the condition without C4-HSL, we listened to Mr. Yihao Zhang’s advice and increased the RBS strength to 60 times of the original one, intending to improve the cooperativity between RhlR and C4-HSL. The figure shows the second device of our sensor. We increased the RBS strength of rhlR (LVA-tagged) to 60 times of the original one by using RBS calculator. We measured the GFP production rate of this device under induction with different concentrations of C4-HSL and modeled the expression pattern by using the Hill equation. The results show that this device can provide a considerable level of fluorescence in fish intestine and feces with the presence of A. hydrophila. The two figures above show our devices for quorum quenching. We used the two autoinducer inactivation enzymes BBa_K2548004 and BBa_K2548005 to replace GFP in the first sensor device. Then, we tested the two devices’ abilities to degrade C4-HSL under induction. The results show that QQ device I can degrade C4-HSL in a higher rate compared with device II (see our demo). The three figures above are the design for the devices to synthesize and extract distinct AMPs in order to test their effectiveness. We chose the pTac promoter, a hybrid of Lac and Trp promoters, to induce AMP production in E. coli because it is the most accessible promoter in our lab. The promoter can be induced by IPTG to remove the repression on transcription from LacI. We also add 6xHis-tag after sequences of three AMPs to extract them out with Nickel column. After testing the antimicrobial abilities of AMPs by using SEM, we intended to construct a device which can release AMPs to fish intestine, which is shown in Fig 8. After the device is induced by C4-HSL, AMPs as well as T4 endolysin can be expressed. The expression of T4 endolysin degrades the peptidoglycan layer of E. coli, so the AMPs can be released and function in fish intestine.The Big Picture
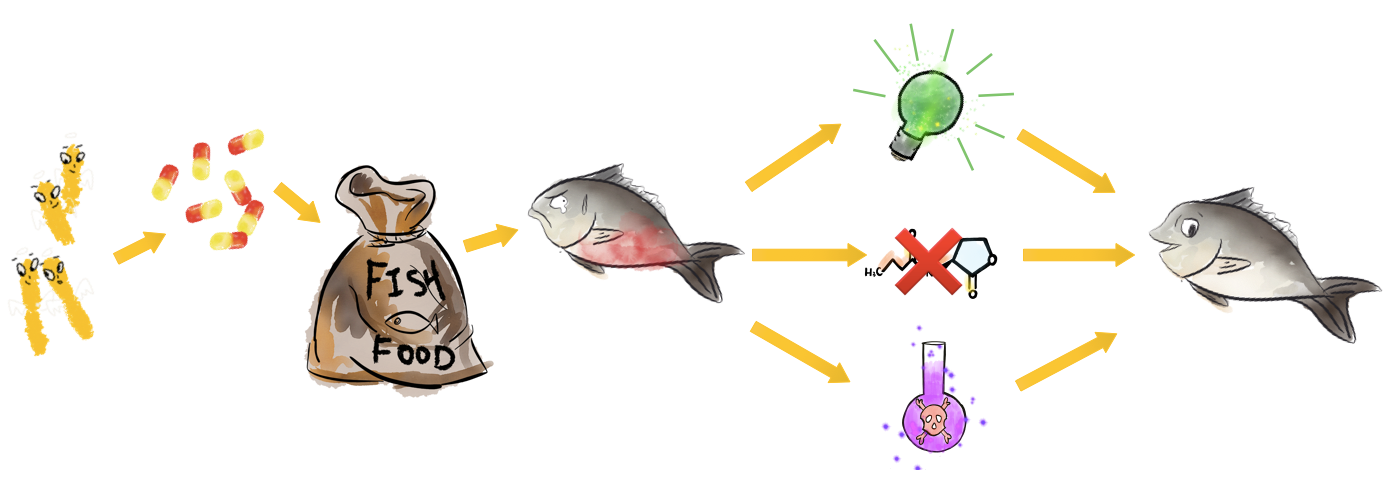
Devices
I. Vector
II. Sensor
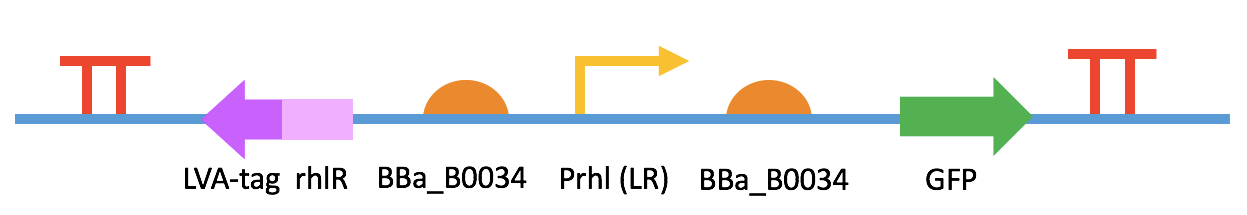
Fig. 1 BBa_K2548000 + BBa_K2548003 (sensor device I) 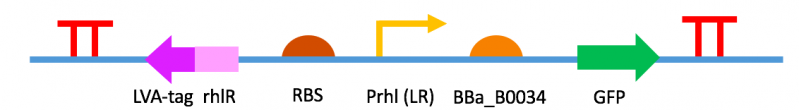
Fig. 2 sensor device II 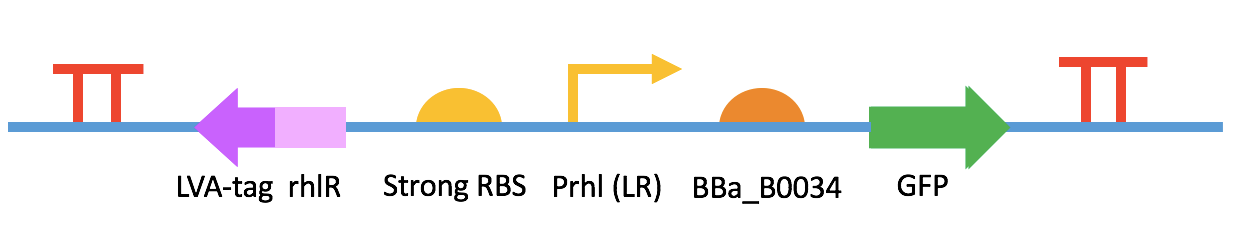
Fig. 3 BBa_K2548001 + BBa_K2548003 (sensor device III) III. Quorum Quenching (QQ)
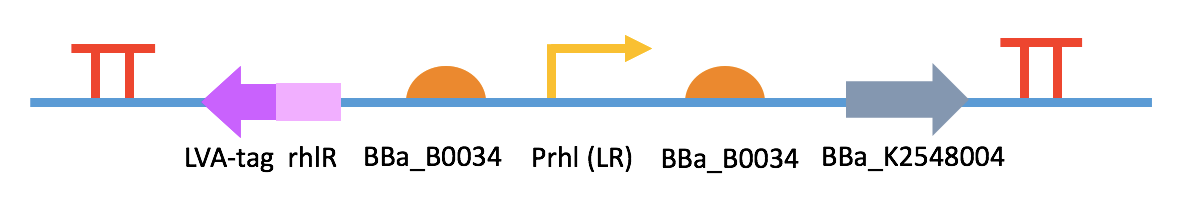
Fig 4. BBa_K2548000 + BBa_K2548006 (QQ device I) 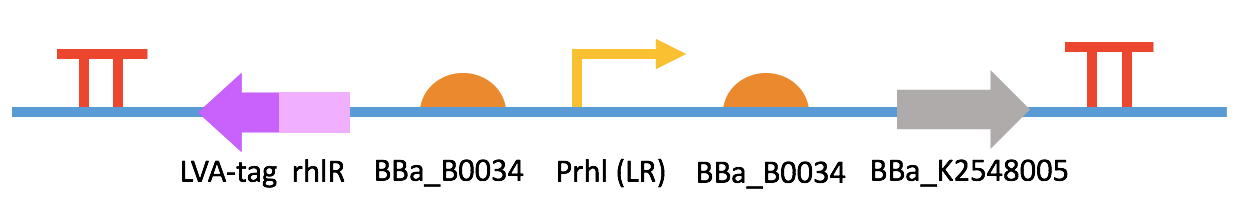
Fig 5. BBa_K2548000 + BBa_K2548007 (QQ device II) IV. Antimicrobial Peptides (AMPs)
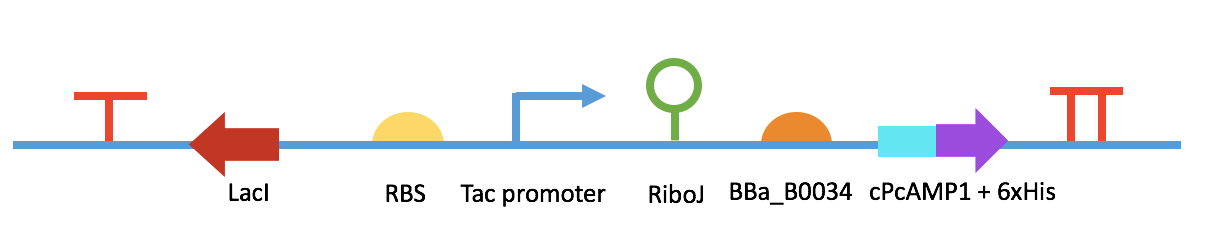
Fig 6. BBa_K2548011 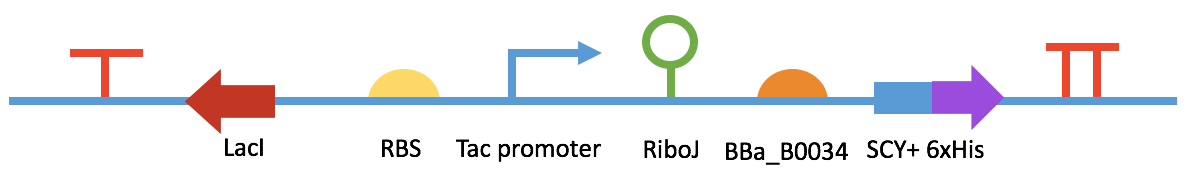
Fig 7. BBa_K2548012 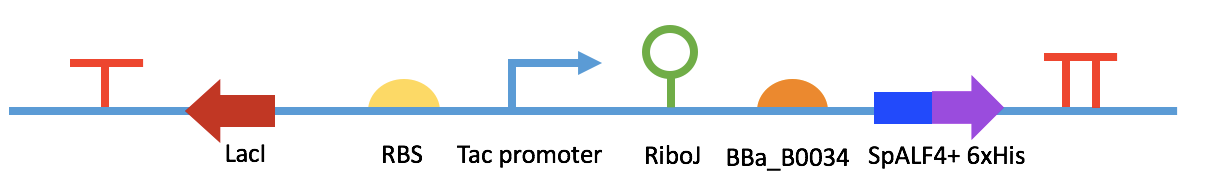
Fig 8. BBa_K2548013 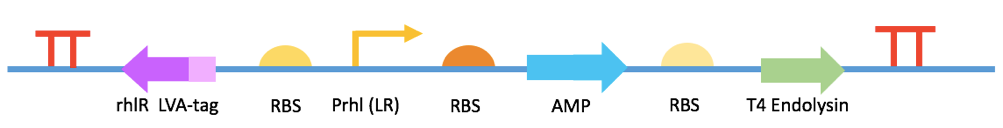
Fig 9. Antimicrobial Peptides Releasing Device
