(Prototype team page) |
|||
| Line 1: | Line 1: | ||
{{BNDS_CHINA}} | {{BNDS_CHINA}} | ||
<html> | <html> | ||
| + | <style> | ||
| + | img { | ||
| + | border: 0mm solid #fdfcf9; | ||
| + | margin: 1px; | ||
| + | box-shadow: 0px 0px 0px rgba(0, 0, 0, 0.3); | ||
| + | } | ||
| + | p {text-indent:2em} | ||
| + | </style> | ||
| + | <body> | ||
| + | fig3: | ||
| + | fig5: | ||
| + | |||
| + | |||
| + | <p>Instead of using AhyR, however, we use RhlR as our transcriptional regulator. RhlR-C4-HSL complex will bind to the promoter region and activate the transcription of the downstream gene in our engineered E. coli.</p> | ||
| + | <div align=center><img width="70%" src="https://static.igem.org/mediawiki/2018/2/20/T--BNDS_CHINA--tani.gif"/> | ||
| + | <p>Fig 3. Quorum Sensing of AhyRI and RhlRI system(就是第一个gif)</p></div> | ||
| + | <p>Many pathogenesis-related factors of A. hydrophila, including expression of virulence factors (such as hemolysin, protease, S layer proteins, DNase, amylase), biofilm maturation, and the type II, III and VI secretion system, are reported under the regulation of quorum sensing in Aeromonas hydrophila. (Cao et al., 2012) In this project, we aim at cutting the QS pathway within A. hydrophila population to cure its infection on fish.</p> | ||
| + | <p>To block QS within A. hydrophila, we engineer E. coli to make it expressing an autoinducer inactivation enzyme aiiA: after the transcriptional regulator RhlR is bound with C4-HSL in the environment, the complex activates the expression of aiiA which inactivate the surrounding C4-HSL to reduce QS within A. hydrophila population. aiiA is a lactonase which cleaves the homoserine lactone ring of molecule AHLs in a hydrolytic and reversible reaction.</p> | ||
| + | <p>Fig 4. Inactivation of C4-HSL</p> | ||
| + | <p>The aiiA we use is AHL-lactonase147-11516 from Bacillus thuringiensis strain 147-11516. It prefers AHLs with shorter acyl chains and thus is observed to have higher specificity with C4-HSL. As it has biological activity under alkaline pH conditions, it is effective in fish’s intestine. (Pedroza, Flórez, Ruiz & Orduz, 2013) We optimize the codons in the sequence BBa_C0160 as the second quorum quenching enzyme.</p> | ||
| + | <div align=center><img width="70%" src="https://static.igem.org/mediawiki/2018/2/2c/T--BNDS_CHINA--description-fig5.gif"/> | ||
| + | <p>Fig 5. Quorum Quenching (第二个gif。。。)</p></div> | ||
| + | |||
| − | + | </body> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</html> | </html> | ||
Revision as of 02:18, 16 October 2018
Instead of using AhyR, however, we use RhlR as our transcriptional regulator. RhlR-C4-HSL complex will bind to the promoter region and activate the transcription of the downstream gene in our engineered E. coli.

Fig 3. Quorum Sensing of AhyRI and RhlRI system(就是第一个gif)
Many pathogenesis-related factors of A. hydrophila, including expression of virulence factors (such as hemolysin, protease, S layer proteins, DNase, amylase), biofilm maturation, and the type II, III and VI secretion system, are reported under the regulation of quorum sensing in Aeromonas hydrophila. (Cao et al., 2012) In this project, we aim at cutting the QS pathway within A. hydrophila population to cure its infection on fish.
To block QS within A. hydrophila, we engineer E. coli to make it expressing an autoinducer inactivation enzyme aiiA: after the transcriptional regulator RhlR is bound with C4-HSL in the environment, the complex activates the expression of aiiA which inactivate the surrounding C4-HSL to reduce QS within A. hydrophila population. aiiA is a lactonase which cleaves the homoserine lactone ring of molecule AHLs in a hydrolytic and reversible reaction.
Fig 4. Inactivation of C4-HSL
The aiiA we use is AHL-lactonase147-11516 from Bacillus thuringiensis strain 147-11516. It prefers AHLs with shorter acyl chains and thus is observed to have higher specificity with C4-HSL. As it has biological activity under alkaline pH conditions, it is effective in fish’s intestine. (Pedroza, Flórez, Ruiz & Orduz, 2013) We optimize the codons in the sequence BBa_C0160 as the second quorum quenching enzyme.
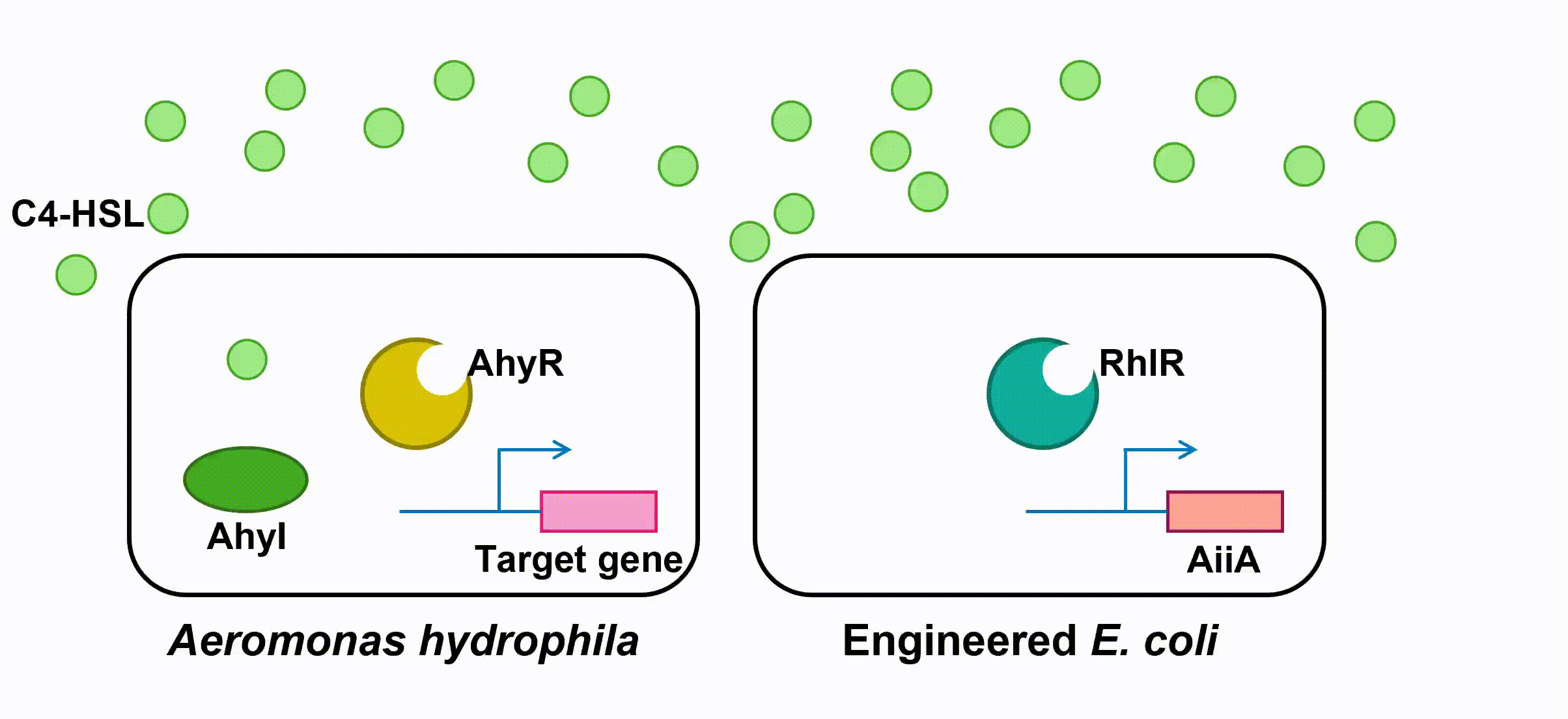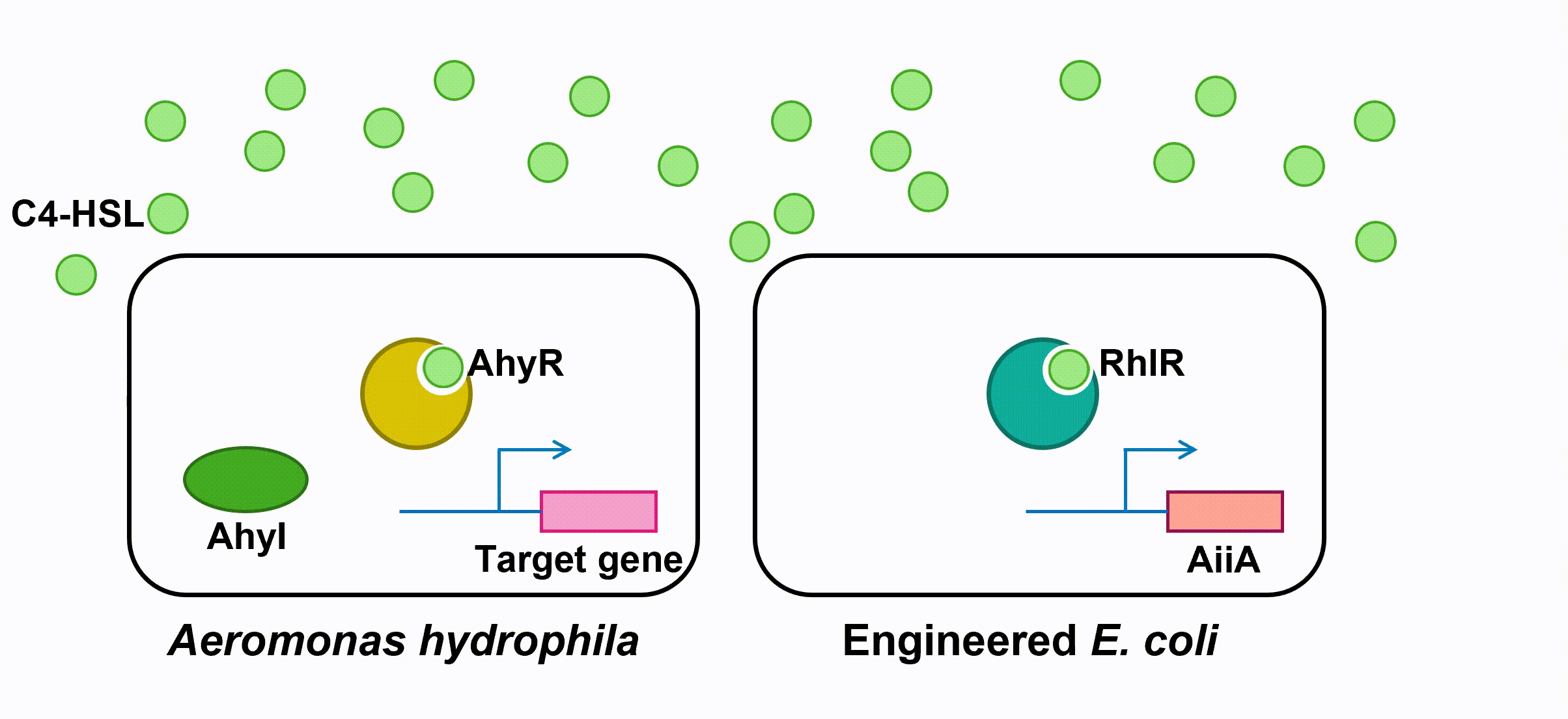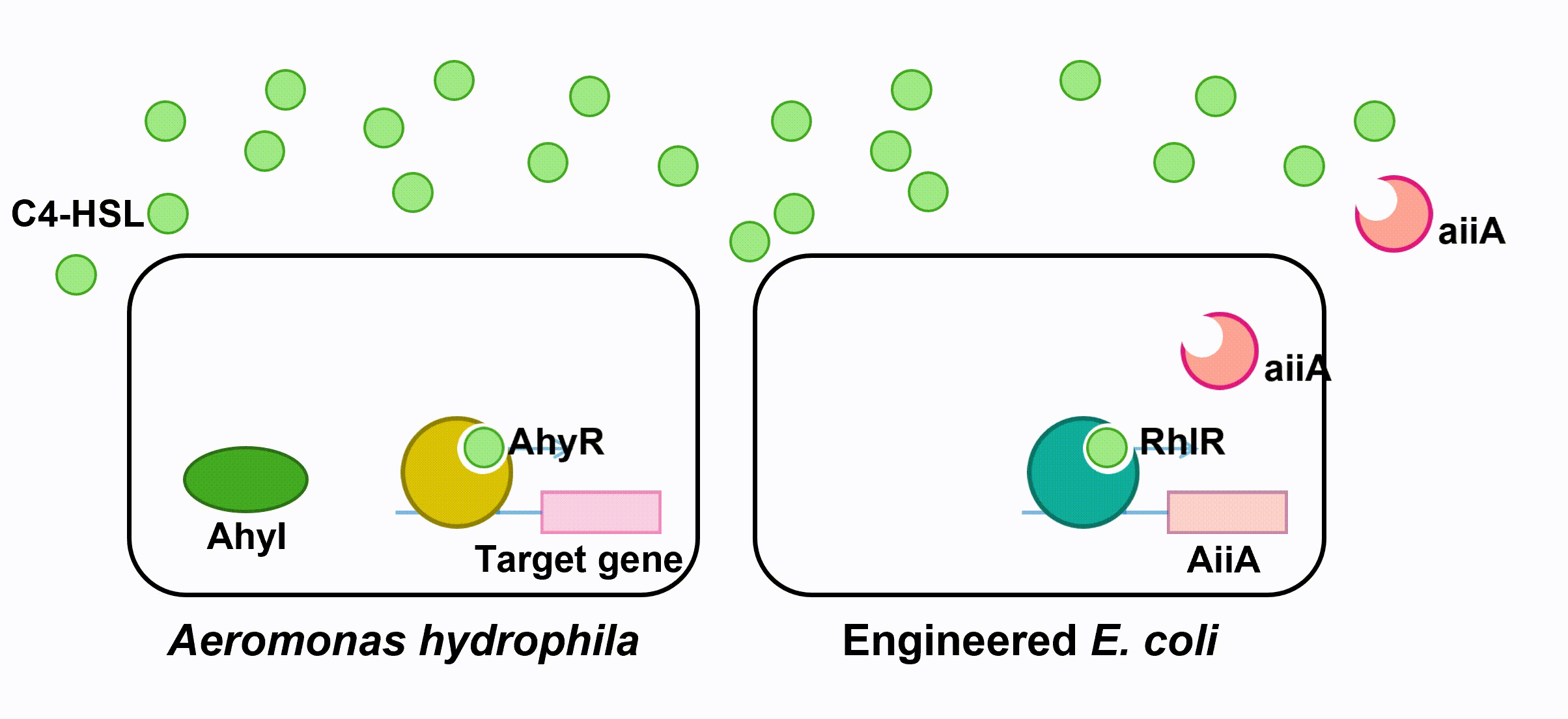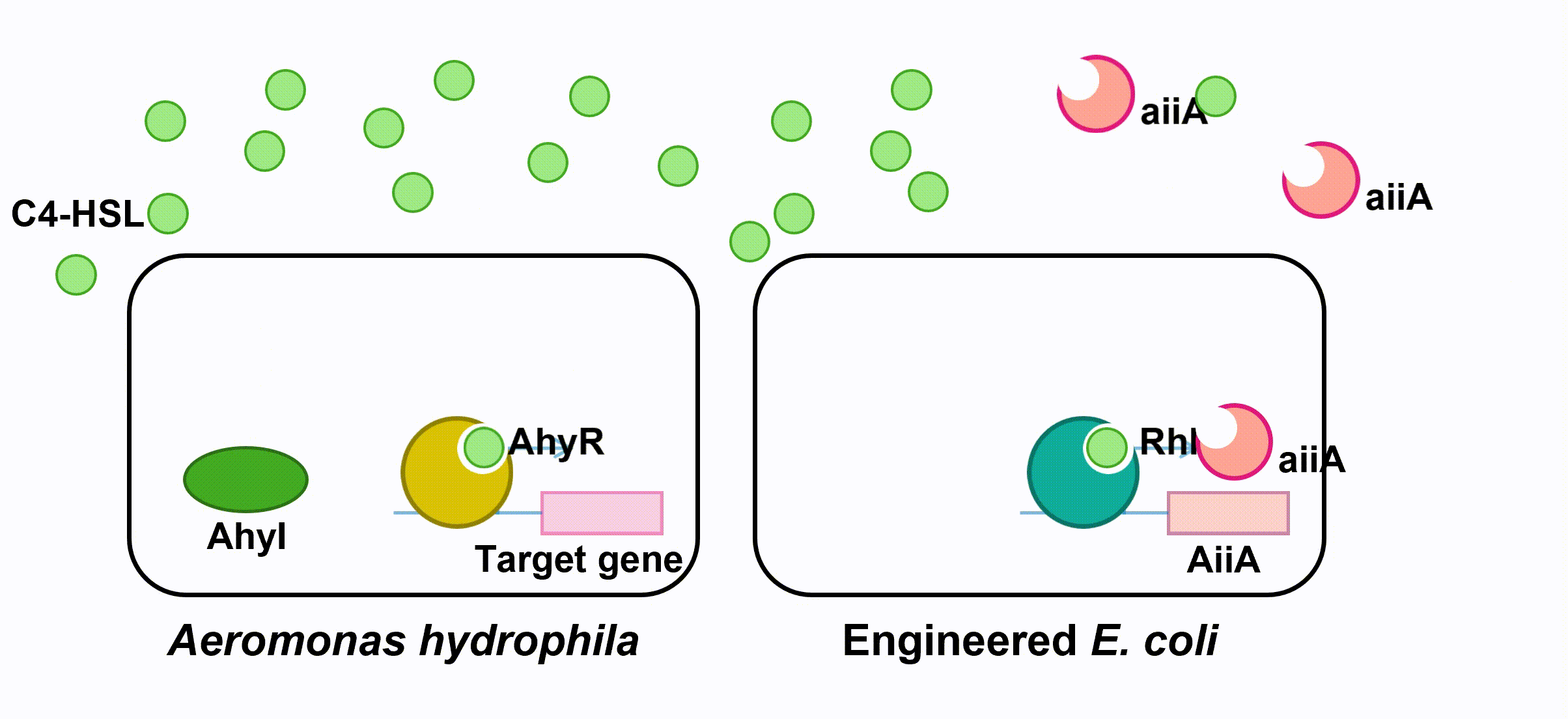
Fig 5. Quorum Quenching (第二个gif。。。)
