| (17 intermediate revisions by 5 users not shown) | |||
| Line 49: | Line 49: | ||
<div class="main-container"> | <div class="main-container"> | ||
| − | <section style=" | + | <section style=" |
| − | + | ; ; background-size: 100%;height: 44.2em;margin-top: 0em;padding-top: 1.1em;" class="height-90 parallax"> | |
| − | + | <div class="main-container" style=" | |
| − | + | height: 100%;"> | |
| − | + | <center style=" | |
| − | + | height: 100%;"> | |
| − | + | <div class="col-md-10" style=" | |
| − | + | height: 100%;margin-top: 4em;"> | |
| − | + | <div id="logoPrinteria" class="item" style=" | |
| − | + | background-image: url(https://static.igem.org/mediawiki/2018/thumb/f/f1/T--Valencia_UPV--ImproveFondoUPV2018.jpeg/1200px-T--Valencia_UPV--ImproveFondoUPV2018.jpeg); | |
| − | + | height: 100%; | |
| − | + | width: 100%;background-attachment: fixed;background-size: cover;"><img src="https://static.igem.org/mediawiki/2018/thumb/d/d7/T--Valencia_UPV--ImproveTextoUPV2018.jpeg/1200px-T--Valencia_UPV--ImproveTextoUPV2018.jpeg.png" style=" margin-top: 0px;"> | |
| − | + | <a class="btn down inner-link active" href="#story" style="font-size: 82%;right: 50%;/* position: fixed; *//* bottom: 7%; */top: 85.2%;z-index: 99;background-color: white;position: absolute;border-radius: 80%;width: 3.8em;height: 3.8em;padding: 0;padding-top: 14px;"> | |
| − | + | <i class="stack-interface stack-down-open-big"></i> | |
| − | + | </a> | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | </center> | |
| − | + | </div><div class="all-page-modals"></div><div class="all-page-modals"></div> | |
| − | + | <div class="all-page-modals"></div> | |
| − | + | </section> | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<a class="anchorOffset" id="story"></a> | <a class="anchorOffset" id="story"></a> | ||
<section class="feature-large" style="padding-top: 6em; padding-bottom: 50px; outline: none;" data-scroll-id="story" tabindex="-1"> | <section class="feature-large" style="padding-top: 6em; padding-bottom: 50px; outline: none;" data-scroll-id="story" tabindex="-1"> | ||
| Line 92: | Line 86: | ||
">Part Improvement</h3> | ">Part Improvement</h3> | ||
<p> | <p> | ||
| − | + | During the development of our iGEM project, particularly when designing our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection" target="_blank">Part Collection</a>, we decided to offer Printeria users a <b>wide range of reporter proteins</b> that they could use in their experiments. However it called our attention that some reporter proteins, such as the <b>Yellow Fluorescent Protein (YFP)</b>, did not present much information about their use and experimentation, compared to the well-characterized Green fluorescent protein (GFP) or (superfolder Green Fluorescent Protein (sfGFP). | |
</p> | </p> | ||
<p> | <p> | ||
| − | + | ||
| + | For this reason, the Printeria team has carried out <b>several experiments using YFP</b> as a reporter protein in order to better characterize it. Throughout our project, we have obtained its <a href="https://2018.igem.org/Team:Valencia_UPV/Experiments#spectra" target="_blank"><b>excitation and emission spectra</b></a>, as well as the molecules of equivalent fluorescence (MEFL) per cell factor that allows us to convert FOD (Fluorescence/OD ratio) into MEFL. | ||
| + | |||
</p> | </p> | ||
<p> | <p> | ||
| − | + | Moreover, we decided to go a step further and improve the YFP coding sequence: <a href="http://parts.igem.org/Part:BBa_K592101"target="_blank"> BBa_K592101</a>. With this aim, we design a new part by adding a LVA ssRA degradation tag (ssRA/tmRNA tag ending in codons coding for leucine, valine and alanine) to the YFP coding sequence, which increases the action of proteases and causes <b>faster degradation</b>. First, we standardized the part to be compatible with the <a href="https://2018.igem.org/Team:Valencia_UPV/Design"target="_blank"> GoldenBraid 3.0</a> assembly method creating the part <a href="http://parts.igem.org/Part:BBa_K2656021"target="_blank"> BBa_K2656021</a>. Then, we added the degradation tag ssRA LVA to the YFP, creating the part <a href="http://parts.igem.org/Part:BBa_K2656020"target="_blank"> BBa_K2656020</a>. | |
</p> | </p> | ||
<p> | <p> | ||
| − | + | ||
| + | To test the effect of the degradation tag on YFP protein amount, we designed an <a href="https://2018.igem.org/Team:Valencia_UPV/Model#tag_exp"target="_blank">experiment</a> to measure the increase in protein degradation due to this tag by comparing two composite parts, with and without the degradation tag. To perform this experiment, we assembled these two composite parts with the same promoter, RBS and terminator: | ||
| + | |||
</p> | </p> | ||
| Line 109: | Line 107: | ||
<li> | <li> | ||
<p> | <p> | ||
| − | <a href="http://parts.igem.org/Part:BBa_K2656112"> BBa_K2656112</a>: | + | <a href="http://parts.igem.org/Part:BBa_K2656112"target="_blank"> BBa_K2656112</a>: |
</p> | </p> | ||
</li> | </li> | ||
<ul style=" | <ul style=" | ||
padding-left: 3em !important;"> | padding-left: 3em !important;"> | ||
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656004">BBa_K2656004</a>: the <a href="http://parts.igem.org/Part:BBa_J23106">J23106</a> promoter in its GoldenBraid compatible version from our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection">Part Collection</a></p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656004"target="_blank">BBa_K2656004</a>: the <a href="http://parts.igem.org/Part:BBa_J23106"target="_blank">J23106</a> promoter in its GoldenBraid compatible version from our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection"target="_blank">Part Collection</a></p></li> |
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656009">BBa_K2656009</a>: the <a href="http://parts.igem.org/Part:BBa_B0030">B0030</a> ribosome biding site in its GoldenBraid compatible version from our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection">Part Collection</a></p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656009"target="_blank">BBa_K2656009</a>: the <a href="http://parts.igem.org/Part:BBa_B0030"target="_blank">B0030</a> ribosome biding site in its GoldenBraid compatible version from our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection"target="_blank">Part Collection</a></p></li> |
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656021">BBa_K2656021</a>: The original part <a href="http://parts.igem.org/Part:BBa_K592101"> BBa_K592101</a> compatible with the GoldenBraid assembly method</p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656021"target="_blank">BBa_K2656021</a>: The original part <a href="http://parts.igem.org/Part:BBa_K592101"target="_blank"> BBa_K592101</a> compatible with the GoldenBraid assembly method</p></li> |
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656026">BBa_K2656026</a>: the <a href="http://parts.igem.org/Part:BBa_B0015">B0015</a> transcriptional terminator in its GoldenBraid compatible version from our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection">Part Collection</a></p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656026"target="_blank">BBa_K2656026</a>: the <a href="http://parts.igem.org/Part:BBa_B0015"target="_blank">B0015</a> transcriptional terminator in its GoldenBraid compatible version from our <a href="https://2018.igem.org/Team:Valencia_UPV/Part_Collection"target="_blank">Part Collection</a></p></li> |
</ul> | </ul> | ||
<li> | <li> | ||
<p> | <p> | ||
| − | <a href="http://parts.igem.org/Part:BBa_K2656111"> BBa_K2656111</a>: | + | <a href="http://parts.igem.org/Part:BBa_K2656111"target="_blank"> BBa_K2656111</a>: |
</p> | </p> | ||
</li> | </li> | ||
<ul style=" padding-left: 3em !important;"> | <ul style=" padding-left: 3em !important;"> | ||
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656004">BBa_K2656004</a></p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656004"target="_blank">BBa_K2656004</a></p></li> |
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656009">BBa_K2656009</a></p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656009"target="_blank">BBa_K2656009</a></p></li> |
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656020">BBa_K2656020</a>: The original part with the added LVA ssRA degradation tag compatible with the GoldenBraid assembly method</p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656020"target="_blank">BBa_K2656020</a>: The original part with the added LVA ssRA degradation tag compatible with the GoldenBraid assembly method</p></li> |
| − | <li><p><a href="http://parts.igem.org/Part:BBa_K2656026">BBa_K2656026</a></p></li> | + | <li><p><a href="http://parts.igem.org/Part:BBa_K2656026"target="_blank">BBa_K2656026</a></p></li> |
</ul> | </ul> | ||
</ul></div> | </ul></div> | ||
| − | <p> Once the experiment was carried out, the results | + | <p> Once the experiment was carried out, from the results in Figure 1 we can observe that the bacteria growth of both constructions is very similar, and the fluorescence (protein amount) had a clear variation.</p> |
| − | <img src="https://static.igem.org/mediawiki/2018/7/72/T--Valencia_UPV--optimization_exp1_YFP_graphUPV2018.png" /> | + | |
| − | + | ||
| + | <a href="https://static.igem.org/mediawiki/2018/7/72/T--Valencia_UPV--optimization_exp1_YFP_graphUPV2018.png" data-lightbox="true"><img src="https://static.igem.org/mediawiki/2018/7/72/T--Valencia_UPV--optimization_exp1_YFP_graphUPV2018.png" alt=""></a></p><h6 style=" | ||
text-align: left !important; | text-align: left !important; | ||
| − | "> Figure 1. | + | "><a href="https://static.igem.org/mediawiki/2018/7/72/T--Valencia_UPV--optimization_exp1_YFP_graphUPV2018.png" data-lightbox="true"></a>Figure 1. Comparison between the transcriptional unit with BBa_K2656021 (YFP) and the one with BBa_K2656020 (YFP LVA tag), and between the experimental and simulation data.</a></h6><a href="https://static.igem.org/mediawiki/2018/7/72/T--Valencia_UPV--optimization_exp1_YFP_graphUPV2018.png" data-lightbox="true"></a> |
| − | <p>These data were optimized | + | |
| + | |||
| + | <p> | ||
| + | |||
| + | These data were optimized using our <a href="https://2018.igem.org/Team:Valencia_UPV/Model#const_models"target="_blank"> model</a>, and the optimized parameters are shown in Table 1. With these parameters it is possible to determine that <b>the protein degradation of the protein with the LVA degradation tag is around twice as much as without the tag</b>. This result shows that the LVA degradation tag improvement can be used for genetic circuits that need a reporter protein with fast degradation.</p> | ||
<table> | <table> | ||
| − | <h6> Table 1. Optimized values of translation | + | <h6>Table 1. Optimized values of translation, Pol degradation and dilution rates from experimental data |
| + | </h6> <tbody><tr> | ||
<th><p>Optimized parameters</p></th> | <th><p>Optimized parameters</p></th> | ||
<th><p>Values</p></th> | <th><p>Values</p></th> | ||
| Line 185: | Line 189: | ||
</tr></tbody></table> | </tr></tbody></table> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 214: | Line 215: | ||
<script type="text/javascript" src="https://2018.igem.org/Template:Valencia_UPV/smoothscrollminJS?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2018.igem.org/Template:Valencia_UPV/smoothscrollminJS?action=raw&ctype=text/javascript"></script> | ||
<script type="text/javascript" src="https://2018.igem.org/Template:Valencia_UPV/jquerystepsminJS?action=raw&ctype=text/javascript"></script> | <script type="text/javascript" src="https://2018.igem.org/Template:Valencia_UPV/jquerystepsminJS?action=raw&ctype=text/javascript"></script> | ||
| − | + | <script type="text/javascript" src="https://2018.igem.org/Template:Valencia_UPV/lightboxminJS?action=raw&ctype=text/javascript"></script> | |
Latest revision as of 01:32, 18 October 2018
Part Improvement
During the development of our iGEM project, particularly when designing our Part Collection, we decided to offer Printeria users a wide range of reporter proteins that they could use in their experiments. However it called our attention that some reporter proteins, such as the Yellow Fluorescent Protein (YFP), did not present much information about their use and experimentation, compared to the well-characterized Green fluorescent protein (GFP) or (superfolder Green Fluorescent Protein (sfGFP).
For this reason, the Printeria team has carried out several experiments using YFP as a reporter protein in order to better characterize it. Throughout our project, we have obtained its excitation and emission spectra, as well as the molecules of equivalent fluorescence (MEFL) per cell factor that allows us to convert FOD (Fluorescence/OD ratio) into MEFL.
Moreover, we decided to go a step further and improve the YFP coding sequence: BBa_K592101. With this aim, we design a new part by adding a LVA ssRA degradation tag (ssRA/tmRNA tag ending in codons coding for leucine, valine and alanine) to the YFP coding sequence, which increases the action of proteases and causes faster degradation. First, we standardized the part to be compatible with the GoldenBraid 3.0 assembly method creating the part BBa_K2656021. Then, we added the degradation tag ssRA LVA to the YFP, creating the part BBa_K2656020.
To test the effect of the degradation tag on YFP protein amount, we designed an experiment to measure the increase in protein degradation due to this tag by comparing two composite parts, with and without the degradation tag. To perform this experiment, we assembled these two composite parts with the same promoter, RBS and terminator:
BBa_K2656004: the J23106 promoter in its GoldenBraid compatible version from our Part Collection
BBa_K2656009: the B0030 ribosome biding site in its GoldenBraid compatible version from our Part Collection
BBa_K2656021: The original part BBa_K592101 compatible with the GoldenBraid assembly method
BBa_K2656026: the B0015 transcriptional terminator in its GoldenBraid compatible version from our Part Collection
BBa_K2656020: The original part with the added LVA ssRA degradation tag compatible with the GoldenBraid assembly method
Once the experiment was carried out, from the results in Figure 1 we can observe that the bacteria growth of both constructions is very similar, and the fluorescence (protein amount) had a clear variation.
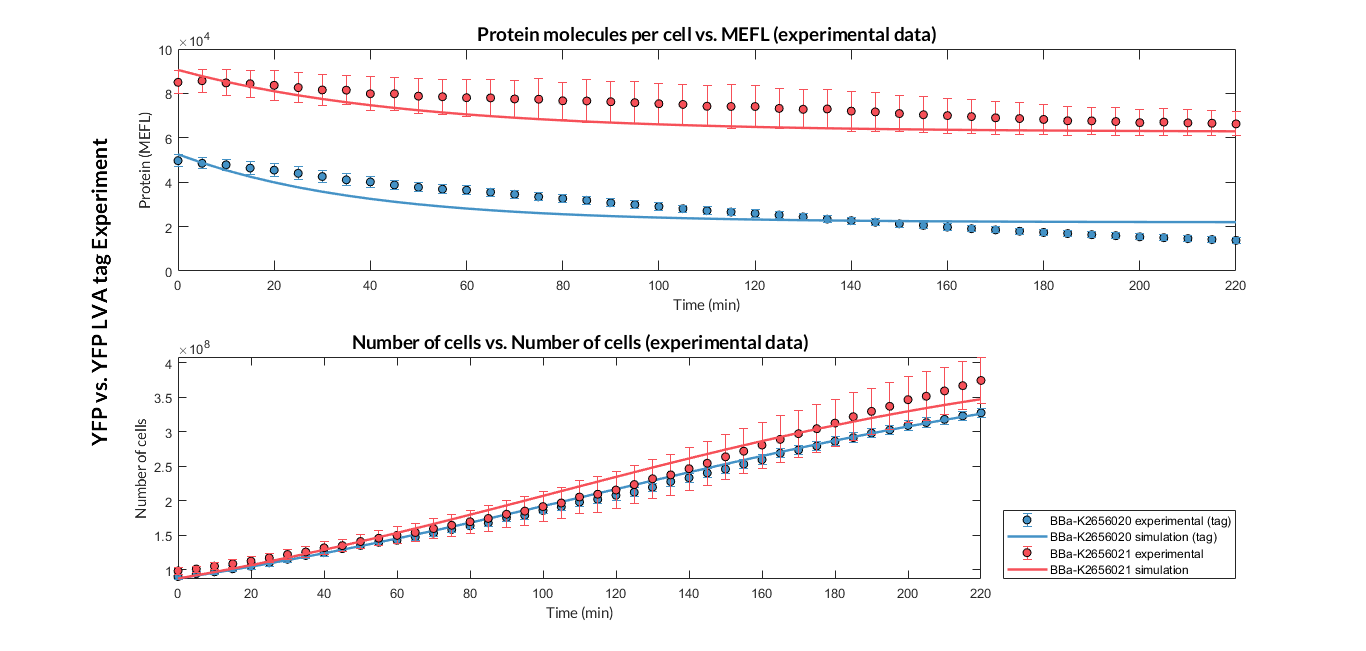
Figure 1. Comparison between the transcriptional unit with BBa_K2656021 (YFP) and the one with BBa_K2656020 (YFP LVA tag), and between the experimental and simulation data.
These data were optimized using our model, and the optimized parameters are shown in Table 1. With these parameters it is possible to determine that the protein degradation of the protein with the LVA degradation tag is around twice as much as without the tag. This result shows that the LVA degradation tag improvement can be used for genetic circuits that need a reporter protein with fast degradation.
Optimized parameters |
Values |
|---|---|
Translation rate p |
|
PoI degradation rate dp |
|
Dilution rate μ |
|


