| Line 2: | Line 2: | ||
==Glycolic acid production in <i>S. cerevisiae</i>== | ==Glycolic acid production in <i>S. cerevisiae</i>== | ||
| − | + | ||
| − | For the production of a biodegradable polymer, our goal was to produce glycolic acid as one of the monomers in <i>Saccharomyces cerevisiae</i>. Glycolic acid production from glyoxylate was initiated by the heterologous expression of glyoxylate reductase (GLYR1). To increase the yield of glycolic acid, we intervened in the glyoxylate cycle by overexpression of the isocitrate lyase (ICL1), by deleting the malate synthase (MLS1) and the isocitrate dehydrogenase (IDP2) (see Koivistoinen et al. CITE ME). We expressed GLYR1 in <i>S. cerevisiae</i>, used Western Blots for verification, purified it via Strep-tag affinity chromatography and characterized its activity through an NADPH assay. | + | {{TU_Darmstadt/Abstract|For the production of a biodegradable polymer, our goal was to produce glycolic acid as one of the monomers in <i>Saccharomyces cerevisiae</i>. Glycolic acid production from glyoxylate was initiated by the heterologous expression of glyoxylate reductase (GLYR1). To increase the yield of glycolic acid, we intervened in the glyoxylate cycle by overexpression of the isocitrate lyase (ICL1), by deleting the malate synthase (MLS1) and the isocitrate dehydrogenase (IDP2) (see Koivistoinen et al. CITE ME). We expressed GLYR1 in <i>S. cerevisiae</i>, used Western Blots for verification, purified it via Strep-tag affinity chromatography and characterized its activity through an NADPH assay.}} |
===Introduction=== | ===Introduction=== | ||
Revision as of 17:37, 6 October 2018
Contents
Glycolic acid production in S. cerevisiae
Abstract
For the production of a biodegradable polymer, our goal was to produce glycolic acid as one of the monomers in Saccharomyces cerevisiae. Glycolic acid production from glyoxylate was initiated by the heterologous expression of glyoxylate reductase (GLYR1). To increase the yield of glycolic acid, we intervened in the glyoxylate cycle by overexpression of the isocitrate lyase (ICL1), by deleting the malate synthase (MLS1) and the isocitrate dehydrogenase (IDP2) (see Koivistoinen et al. CITE ME). We expressed GLYR1 in S. cerevisiae, used Western Blots for verification, purified it via Strep-tag affinity chromatography and characterized its activity through an NADPH assay.
Introduction
By modifying the natural glyoxylate cycle, we aim to generate a biosynthetic pathway to produce glycolic acid in S. cerevisiae using the strains CEN.PK-1C and H3847 (HERKUNFT- Link zur Finnland Quelle).
Bild pathway in s. cerevisiae/ glyoxylate cycle
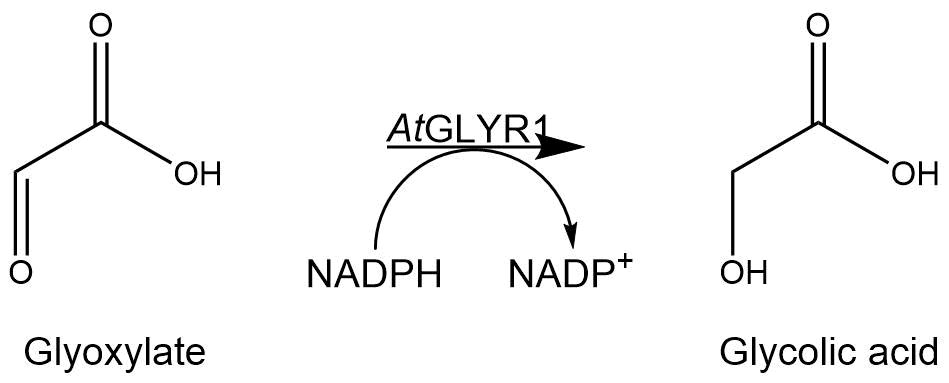
GOR1 is the natural occurring gene in S. cerevisiae, which encodes the enzyme converting glyoxylate into glycolic acid by reducing the aldehyde group in a NADPH-dependent reaction. Since the gene is not expressed (CITE https://onlinelibrary.wiley.com/doi/epdf/10.1002/yea.1434) under normal growth conditions and the encoded glyoxylate reductase shows moderate affinity to the substrate glyoxylate, we used GLYR1 from Arabidopsis thaliana instead. Previous studies reported an enhanced affinity of AtGLYR1 to glyoxylate (CITE http://www.nrcresearchpress.com/doi/abs/10.1139/B07-081#.W7CnJhMzYk8) and also enhanced activity (CITE Koivistoinen et al.). The enzyme has a molecular weight of 30,7 kDa (https://www.uniprot.org/uniprot/Q9LSV0).
Accumulating glyoxylate benefits the yield of glycolic acid. The isocitrate lyase 1, ICL1, catalyzes the reaction from isocitrate to glyoxylate. In the formation of glyoxylate, succinate occurs as a byproduct of the reaction. By overexpressing ICL1, a high substrate concentration is ensured. ICL1 has a molecular weight of 62.4 kDa (https://www.yeastgenome.org/locus/S000000867#protein)
The accumulation of glyoxylate is supported by the deletion of MLS1 and IDP2, since these enzymes degrade glyoxylate and isocitrate in unwanted side reactions. Consequently, previous deletions of both enzymes in S. cerevisiae significantly increased glycolic acid production titers (CITE Koivistoinen). MLS1, a malate synthase, catalyzes the conversion from glyoxylate to malate, and IDP2, an isocitrate dehydrogenase, catalyzes the oxidation of isocitrate to α-ketoglutarate. As a result of our metabolic engineering approach, we were able to measure glycolic acid production in S. cerevisiae.
Methods
Cloning
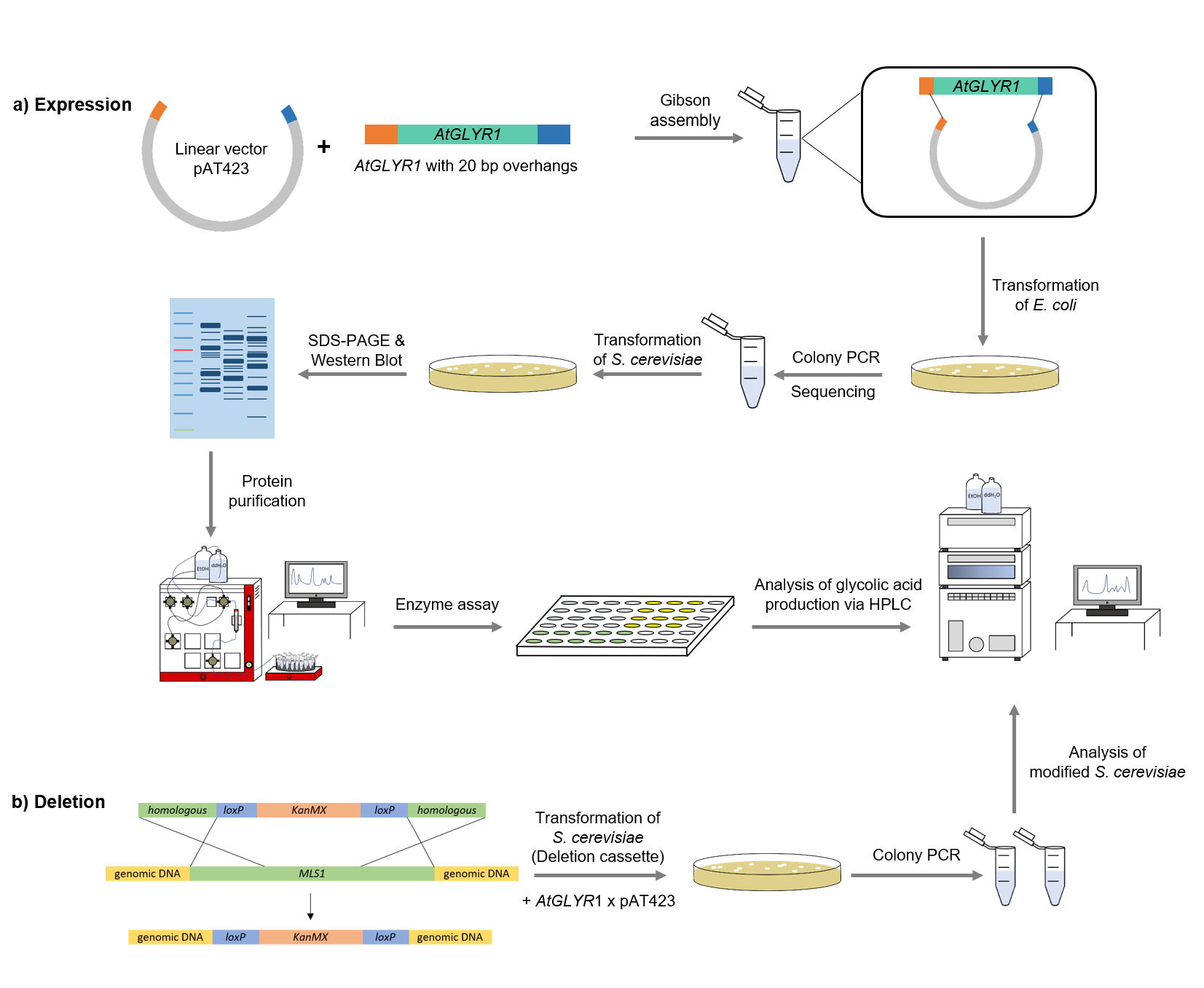
The sequence for AtGLYR1 was firstly modified with a Strep-tag on the 3´end and 20 base pairs overhangs at the two ends of the gen, which are complementary with the alcohol dehydrogenase promotor (ADH) of the pAT423 plasmid. The sequence was ordered from Integrated DNA Technologies (IDT) and inserted into the pAT423 plasmid using the Gibson assembly method. E. coli TOP10 were transformed with generated plasmids and positive colonies were identified via colony PCR and DNA sequencing.
 [[->2 Plasmidkarten pro Teamseite]]
[[->2 Plasmidkarten pro Teamseite]]
Deletion
MLS1 and IDP1 were deleted of the genome of the S. cerevisiae using deletion cassettes, which were integrated in the genome by homologous recombination. The both deletion cassettes (KanMX-cassette and Leucin-cassette) were firstly amplified using primers, which bound at the loxP-sides and had 40 bp homolog to the gene, using PCR. After that, S. cerevisiae were transformed with the linearized amplificated deletion cassettes, so that MLS1 was deleted with the KanMX- and IDP2 with the Leucin-cassette.
SDS-PAGE and Western Blot
To verify that AtGLYR1 was expressed and the corresponding protein was translated, a SDS-PAGE and Western Blot were performed. The resulting bands were compared to the expected protein sizes of 32,9 kDa.
Purification
After expression of AtGLYR1 in S. cerevisiae, an ÄKTA system was used to purify the desired Strep/His-tagged enzyme(s).
Enzyme assays
The assay for the glyoxylate reductase AtGLYR1 is based on the different absorption maxima of NADPH (absorption maximum at 340 nm) and NADP (maximum at 260 nm). During the reaction the enzyme uses NADPH as a cofactor. NADPH is converted into NADP, which leads to a decrease of absorption at 340 nm. By measuring the absorption level over time, it is possible to quantify the enzyme activity and glycolic acid production.
HPLC analysis
To detect the precursors, as well as the produced monomer glycolic acid, high-performance liquid chromatography (HPLC) was employed. An organic acid separation column as stationary phase and sulfuric acid as mobile phase allowed the separation of isocitrate, glyoxylate and glycolic acid. Signals were recorded by a refractive index detector.
Results
Discussion and Outlook