Modeling
Do you think it is possible to mathematically describe a cell? Would you like to know the possibilities that Printeria modeling offers you?
One of the fundamental bases of the Printeria project has undoubtedly been mathematical modeling. Thanks to the development and application of new mathematical models, it is possible to quantify the expression of proteins in cells, and therefore characterize through different experiments the parts designed by Printeria. From the Printeria modeling team, we intend to reach different goals:
Design simple mathematical models based on differential equations that describe the biochemical processes of a cell. With them, we can simulate the different genetic circuits that Printeria allows us to build.
Develop a Simulation Tool that allows the user to visualize a prediction of the results of their experiment before running it in Printeria.
Optimize model parameters to match simulation results to experimental data obtained from Printeria constructions.
Characterize the parts of our Part Collection from the optimization results and provide the user with all the information about the Printeria kit.
Although in the development of the project we have dealt with all these aspects, all of them have a single purpose: to understand and describe in a mathematical way the biological processes that take place inside the cell.
Models & Experiments
Characterisation procedures
Valencia UPV iGEM team has designed an extensive Part Collection in purpose of allowing the user to design multiple genetic constructions and experiments. One of our main objectives has been to show the user clear and structured information about the pieces that make up the Printeria kit. For this reason, we have considered the characterization of the parts as a priority when developing the project. In this way, we have elaborated some procedures which have allowed us to systematically obtain and structure information from the parts.
Procedure for obtaining protein spectra
Obtaining excitation and emission spectra is a fundamental aspect in the process of characterization of a fluorescent protein. Each protein has a characteristic spectrum, which indicates the energy in which the molecule is excited or emits at a certain wavelength.
The characterization of the reporter proteins by excitation and emission spectra are of great importance in the experimentation. When contrasting experimental information with the theoretical results of mathematical models, we experiment with numerous reporter proteins, such as fluorescence proteins or chromoproteins. However, the >fluorescence data obtained must be corrected applying diferent operations in order to obtain representative fluorescence data:
The subtraction of the blank fluorescence:
F_{subs} = F - F_{blank}The quotient of the fluorescence with the gain:
F_{gain} = \frac{F_{subs}}{G}The quotient of the fluorescence with excitation and emission efficiency:
F_{corrected} = \frac{F_{gain}}{\frac{exc(\%)em(\%)}{100}}

Until now, the corrections applied to the experiments performed with these reporter proteins were the subtraction of the fluorescence of the blank, and the division by the gain factor of the measuring equipment. However, with the protein spectra, we can also normalize the fluorescence data to values that would have been obtained with maximum excitation and emission.
Owing to this reason, a protocol has been established in the lab by Lab and Modeling team to obtain the spectrum of any reporter protein.
Materials:
-
Measuring equipment: Biotek Cytation3
-
96 well plate
-
MATLAB 2018a software
Procedure:
We look for the theoretical spectraof the protein to be measured (or similar molecules) to determine the wavelength at which the protein is excited or emitted at maximum energy, i.e. where the theoretical spectral peaks occur.
We define the protocol of our equipment to get the absorbance and fluorescence dataset. In our protocol, the most important parameters to be established are summarized in the following Table.
Parameters
Description
Number of samples
From 3 to 6 samples of reporter protein and 3 samples of blank (BLK)
Temperature
37 ºC
Shake
Double Orbital. 01:00 (MM:SS)
Absorbance. Optical Density (OD)
Wavelenght at 600 nm emission
Excitacion and emission scans
The scans occur between two wavelength limit values. The established range will depend on the theoretical spectrum of the protein.
Excitacion and emission wavelengths
These values will depend on the range, and therefore on the spectrum. Values far from the theoretical peak lead to more attenuated fluorescence curves, and values very close to the peak can lead to overlap and error in reading the data. Therefore, a compromise must be reached between curve resolution and reading overlap.
Gain (G)
Normally the gain value G = 60, although for proteins with lower fluorescence, it is recommended that G takes higher values.
The experiment is introduced, and the experimental absorbance data and the fluorescence curves of the samples are obtained with the blank correction applied.
The dataset is exported to an Excel file.
The MATLAB script spectrum.m for fluorescent proteins is executed:
-
Dataset is extracted from the Excel file.
-
The fluorescence curves of all samples are averaged, and the result is normalised (from 0 to 100%).
-
Graphs of the normalized absorption and emission spectra are plotted. The X-axis represents the wavelength (nm), and the Y-axis represents the normalized fluorescence intensity (%).
-
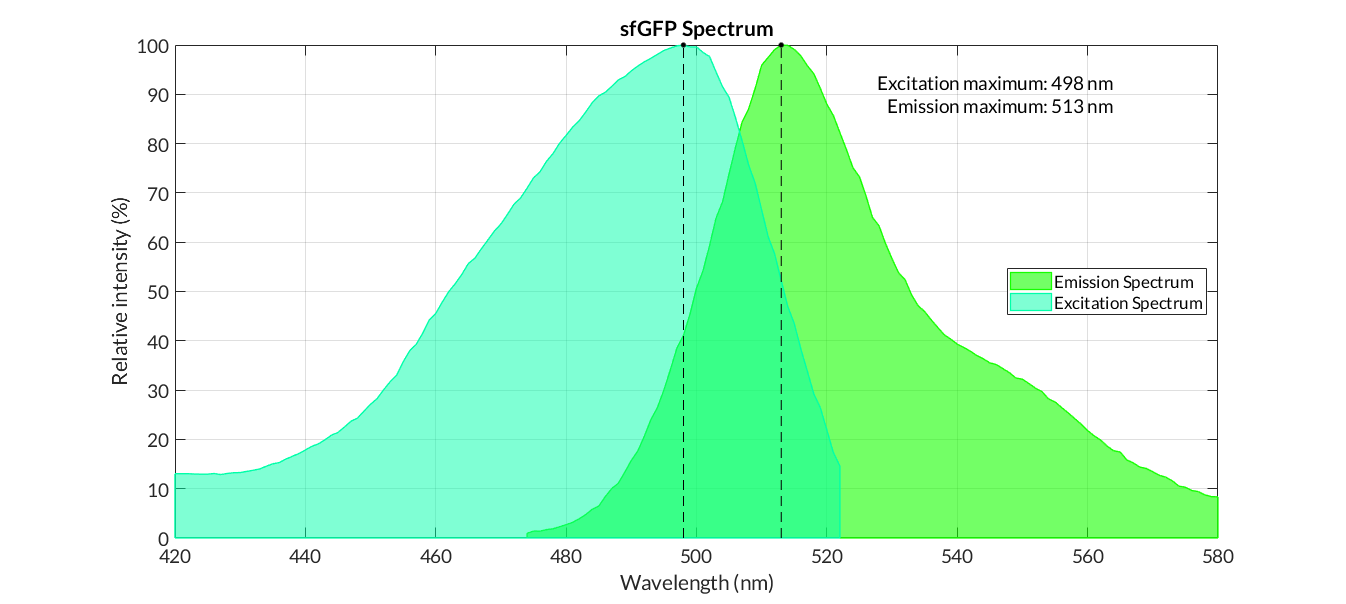
sfGFP spectra
>In practical terms, the protocol has been applied to all of our reporter proteins: GFP, sfGFP, YFP and mRFP. All results can be found in our parts collection as well as in the iGEM catalogue.
In the particular case of a reporter chromoprotein, such as amilCP, we do not measure fluorescence, but absorbance. In this case, in rder to obtain the corrected absorbance curve, we must subtract from the cell absorbance data with the reporter protein the absorbance of a medium with cells without chromoprotein. Once the data have been corrected, we normalize them between values of 0 and 100 and with this we elaborate the graph. The protocol used can be found in the MATLAB script amilCP_spectrum.m.
Finally, we have also obtained the spectra of the fluorescein molecule. These spectra have been used to correct the fluorescence data used in the Interlab Study to obtain the Relative Fluorescence Units (RFU) to Molecules of Equivalent Fluorochrome (MEFL) conversion factor. In addition, the comparison between GFP and sfGFP proteins RFU has been possible thanks to fluorescein spectra.

Fluorescein spectra
>Comparison between sfGFP and GFP relative fluorescence intensity
One of the main problems we encounter when processing the results of experiments in Synthetic Biology are the units of measurement of fluorescence data. Unlike absorbance, where there is a simple conversion between Optical Density (OD) and cell number, there is no direct relationship between the Relative Units of Fluorescence (RFU) and the number of protein molecules in the cell. Moreover, RFUs vary among reporter fluoroproteins: for example, an RFU of the GFP protein does not have to be equivalent to an RFU of the mRFP protein, sfGFP, etc.
Thanks to initiatives such as the Interlab Study, we have been able to go a step further and obtain a MEFL/RFUGFP factor of equivalence between the RFU of the GFP protein and the Molecules of Equivalent Fluorochrome (MEFL). This relationship is an important breakthrough, as it can give us a more accurate estimate of the amount of GFP molecules in the cell.
However, another of the reporter proteins widely used in the experimentation is the sfGFP. This protein has a much faster folding than GFP, which translates into a higher fluorescence intensity per molecule. In order to obtain the MEFL/RFUsfGFP factor from sfGFP, the Printeria Modeling and Lab teams have designed a comparative experiment between the both proteins. The experiment consists of designing two identical transcriptional units (TU), changing only the CDS sequence so that each TU will produce GFP and sfGFP, respectively. It should be added that this experiment is based on two fundamental assumptions:
The number of GFP molecules produced in the cells is equivalent to the number of MEFL.
Given two TU with identical promoters, RBS and terminators, but with different CDS, under the same experimental conditions, the number of molecules produced by each TU is the same.
Taking these axioms into account, the materials and procedure followed to calculate the MEFL/RFUsfGFPfactor were as follows:
Materials:
-
Measuring equipment: Biotek Cytation3
-
96 well plate
-
MATLAB 2018a software
Procedure:
From the fluorescein spectrum, and the fluorescence data obtained from the Interlab Study experiment, blank, gain and efficiency corrections have been made in the data set.
The data are introduced in the Excel file of the Interlab Study. With them we obtain the factor MEFL/RFUGFP. In our case, MEFL/RFUGFP = 271.2 (GFP equivalent molecules/RFU)
We establish the protocol of the experiment and the parameters of our equipment. The experiment consists in the measurement of the absorbance and fluorescence of two TU of identical structure, whose CDS codifies for GFP and sfGFP proteins. The most relevant information of the experiment is detailed in the following Table:
Parameters
Description
Time
06:00:00 (HH:MM:SS). Measurement interval: 05:00 (MM:SS)
Number of samples
8 samples for each TU. Total samples: 16
Temperature
37 ºC
Shake
Double Orbital Continuous
Absorbance. Optical Density (OD)
Wavelenght at 600 nm emission
Excitacion wavelength
485 nm
Emission wavelength
528 nm
Gain
60
The experiment is introduced into the equipment and the absorbance and fluorescence curves are obtained, as well as the curve of {Fluorescence F/Absorbance OD} (FOD) ratio with the applied blank correction.
The absorbance and fluorescence data are exported to an Excel file.
Running the MATLAB GFP_sfGFP_comparison.m script:
-
We extract the FOD data from the GFP and sfGFP from the Excel file.
-
We apply the gain and efficiency corrections of the spectrum to the FOD.
-
We plot the FOD curves and look for a stationary equilibrium time interval in the expression of the GFP and sfGFP proteins. In our experiment, we have decided to chose the interval [145,290] min
-
We obtain the average value of both regions.
-
We calculate the number of GFP MEFLs by multiplying the FOD data by the MEFL/RFUGFP factor.
-
If we assume that the number of molecules expressed by both TU is the same, the number of MEFL for the calculated GFP is the same as the number of MEFL for the sfGFP.
-
We calculate the MEFL/RFUsfGFP factor by dividing the MEFL number by the average FOD value of the sfGFP in the stationary region. In our case, MEFL/RFU
sfGFP= 121.4.
-

sfGFP and GFP FOD (a.u) vs Time (min)
>Reporter protein |
MEFL/RFU factor |
|---|---|
GFP |
273.7 |
sfGFP |
121.4 |
With MEFL/RFU factors, FOD data obtained in any experiment in which GFP or sfGFP was used as reporter proteins can be transformed into equivalent fluorescein molecules by applying the following ratio: Molecules = (MEFL/RFU) x FOD.
The new data give us a more accurate estimate of the number of molecules in the cell. Consequently, by relating the experimental results with the theoretical mathematical models in the optimization process, the parameters of the model acquire values more consistent with their physical significance, working in equivalent molecules and not in arbitrary units.
Simulation Tool


