PeterGockel (Talk | contribs) |
|||
| Line 5: | Line 5: | ||
===Glycolic acid production in <i>E. coli </i>=== | ===Glycolic acid production in <i>E. coli </i>=== | ||
| − | Our aim was to produce glycolic acid for the production of biodegradable polymers. We were able to overexpress the genes <i>aceA</i> and <i>ycdW</i> in Escherichia coli. The proteins AceA and YcdW were tagged with | + | Our aim was to produce glycolic acid for the production of biodegradable polymers. We were able to overexpress the genes <i>aceA</i> and <i>ycdW</i> in Escherichia coli. The proteins AceA and YcdW were tagged with a His-tag and purified via an ÄKTA system. The purification was confirmed by an SDS-PAGE and western blot. After that we successfully characterized YcdW with an NADPH dependent enzyme assay. The assay indicates that YcdW uses glyoxylate as substrate under consumption of NADPH to produce glycolic acid. The functionality of AceA was verified with an phenylhydrazine-dependent assay. AceA catalyzes the reaction from isocitrat to glyoxylate. All performed assay were furthermore analyzed via HPLC. |
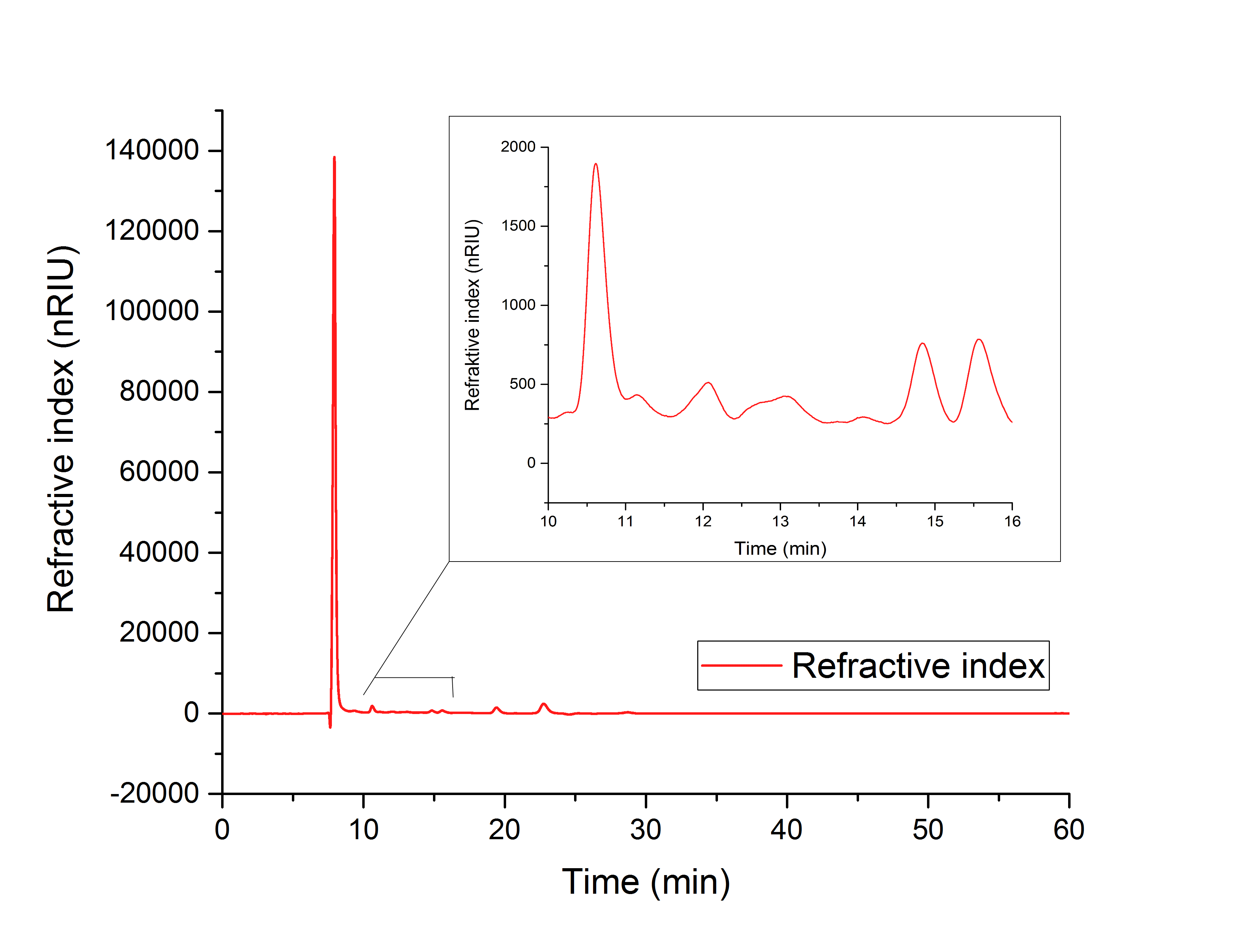
[[image:T--TU_Darmstadt--hplcvivo.jpg|500px |alt=Text|HPLC of disrupted ycdW transformed <i>E. coli</I> cells]] | [[image:T--TU_Darmstadt--hplcvivo.jpg|500px |alt=Text|HPLC of disrupted ycdW transformed <i>E. coli</I> cells]] | ||
| + | |||
| + | In each one of them, the expected product was found, indicating a catalytic activity of the purified enzymes. When the enzyme YcdW was expressed in <i>E.Coli</i>, the formation of glycolic acid was observed in the disrupted cells (Figure XY). The combined evidence shows, that the transformation of <i>E.Coli</i> was successful, leading to the expression of functioning enzymes. These enzymes then partake in the production pathway of glycolic acid, which was shown to function in vivo as well as in vitro. | ||
===Glycolic acid production in <i>S. cerivisiae</i>=== | ===Glycolic acid production in <i>S. cerivisiae</i>=== | ||
Revision as of 10:27, 13 October 2018
Proof of Concept
Glycolic acid production in E. coli
Our aim was to produce glycolic acid for the production of biodegradable polymers. We were able to overexpress the genes aceA and ycdW in Escherichia coli. The proteins AceA and YcdW were tagged with a His-tag and purified via an ÄKTA system. The purification was confirmed by an SDS-PAGE and western blot. After that we successfully characterized YcdW with an NADPH dependent enzyme assay. The assay indicates that YcdW uses glyoxylate as substrate under consumption of NADPH to produce glycolic acid. The functionality of AceA was verified with an phenylhydrazine-dependent assay. AceA catalyzes the reaction from isocitrat to glyoxylate. All performed assay were furthermore analyzed via HPLC.
In each one of them, the expected product was found, indicating a catalytic activity of the purified enzymes. When the enzyme YcdW was expressed in E.Coli, the formation of glycolic acid was observed in the disrupted cells (Figure XY). The combined evidence shows, that the transformation of E.Coli was successful, leading to the expression of functioning enzymes. These enzymes then partake in the production pathway of glycolic acid, which was shown to function in vivo as well as in vitro.
Glycolic acid production in S. cerivisiae
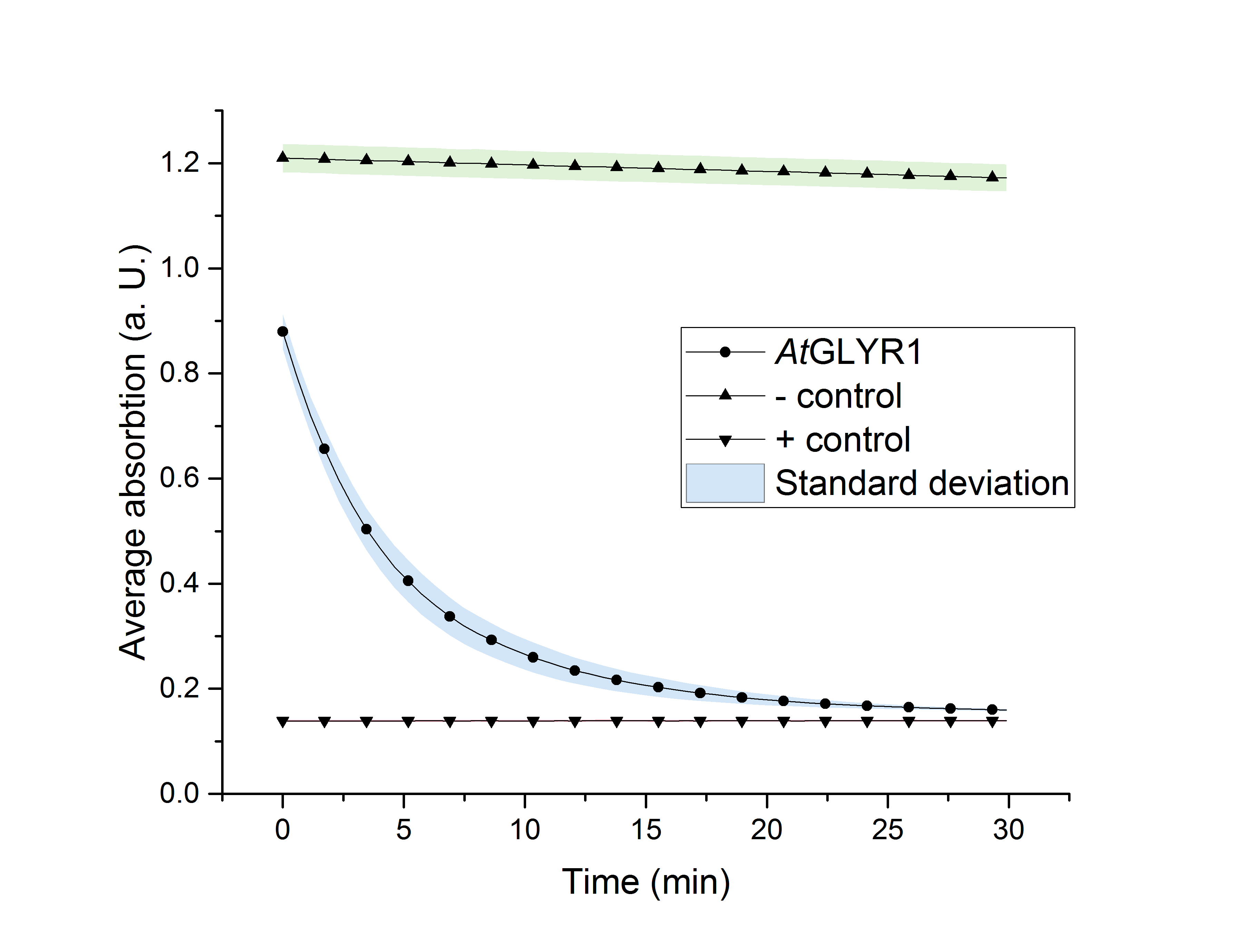
As glycolic acid is one monomer which is needed to produce PLGA our goal was to produce glycolic acid in S. cerevisiae. It was possible for us to successfully express AtGLYR1 in S. cerevisiae. This was shown by western blot analysis as well as during an enzyme assay. Using a NADPH dependent enzyme assay we could proof the functionality of our purified enzyme. This is shown in Figure X. During the assay AtGLYR1 converts glyoxylate to glycolic acid while turning over NADPH. Therefore, we were able to produce glycolic acid in vitro. Furthermore, we were able to delete several genes from the genome of CEN.PK 1C. This could be helpful to increase the production of glycolic acid in vivo.