| Line 22: | Line 22: | ||
====Mechanism==== | ====Mechanism==== | ||
===Methods=== | ===Methods=== | ||
| + | [[image:T--TU_Darmstadt--E.coli,Methodenteil.jpg|100px|alt=Text|E.coli Methoden]] | ||
| + | |||
| + | |||
| + | |||
====Cloning==== | ====Cloning==== | ||
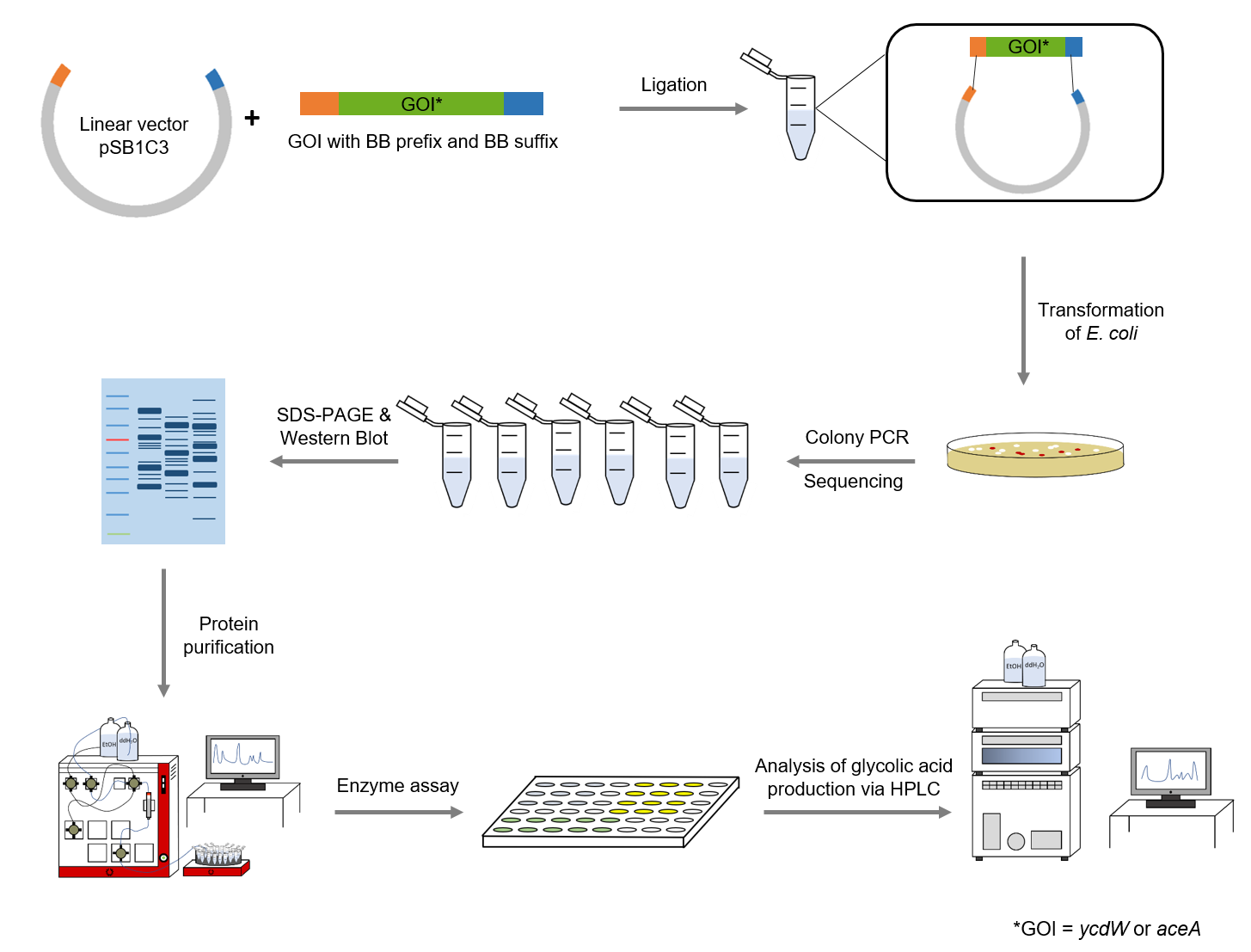
The sequences for aceA and ycdW were modified with a His-tag, ordered from Integrated DNA Technologies (IDT) and inserted into the pSB1C3 plasmid. For this purpose, the BioBrick assembly (BBa) was used. A T7 lac promotor (BBa_K921001) and a B0034-based ribosomal binding site (BBa_K2380024) were inserted upstream of the coding sequence via the BBa as well. <i>E. coli</i> TOP10 were transformed with generated plasmids and positive colonies were identified via colony PCR and DNA sequencing. | The sequences for aceA and ycdW were modified with a His-tag, ordered from Integrated DNA Technologies (IDT) and inserted into the pSB1C3 plasmid. For this purpose, the BioBrick assembly (BBa) was used. A T7 lac promotor (BBa_K921001) and a B0034-based ribosomal binding site (BBa_K2380024) were inserted upstream of the coding sequence via the BBa as well. <i>E. coli</i> TOP10 were transformed with generated plasmids and positive colonies were identified via colony PCR and DNA sequencing. | ||
Revision as of 12:55, 5 October 2018
Contents
Glycolic acid production in E. coli
Abstract
Our aim was to produce glycolic acid for the production of biodegradable polymers. Hence, we overexpressed the enzymes isocitrate lyase (AceA) and glyoxylate reductase (YcdW) in Escherichia coli (E. coli) to synthesized glycolic acid from the TCA cycle intermediate isocitrate. In the end we successfully characterized our enzymes and verified the production of glycolic acid through both, NADPH and phenylhydrazine-dependent assays, as well as HPLC analysis. We therefore created a pair of BioBricks allowing the community to produce glycolic acid, and submitted them to the iGEM Registry (LINK ZU PART NO.).
Introduction
Escherichia coli is the most frequently used and a well-established host organism for biotechnological applications. Especially its stable expression system of heterologous genes, fast growth rates and its overall well characterized metabolome and proteome make it a currently indispensable organism in biotechnology.
In our project, we modified the glyoxylate cycle of E. coli to produce glycolic acid. For this purpose, we overexpressed two native E. coli genes encoding the isocitrate lyase AceA and the glyoxylate reductase YcdW.
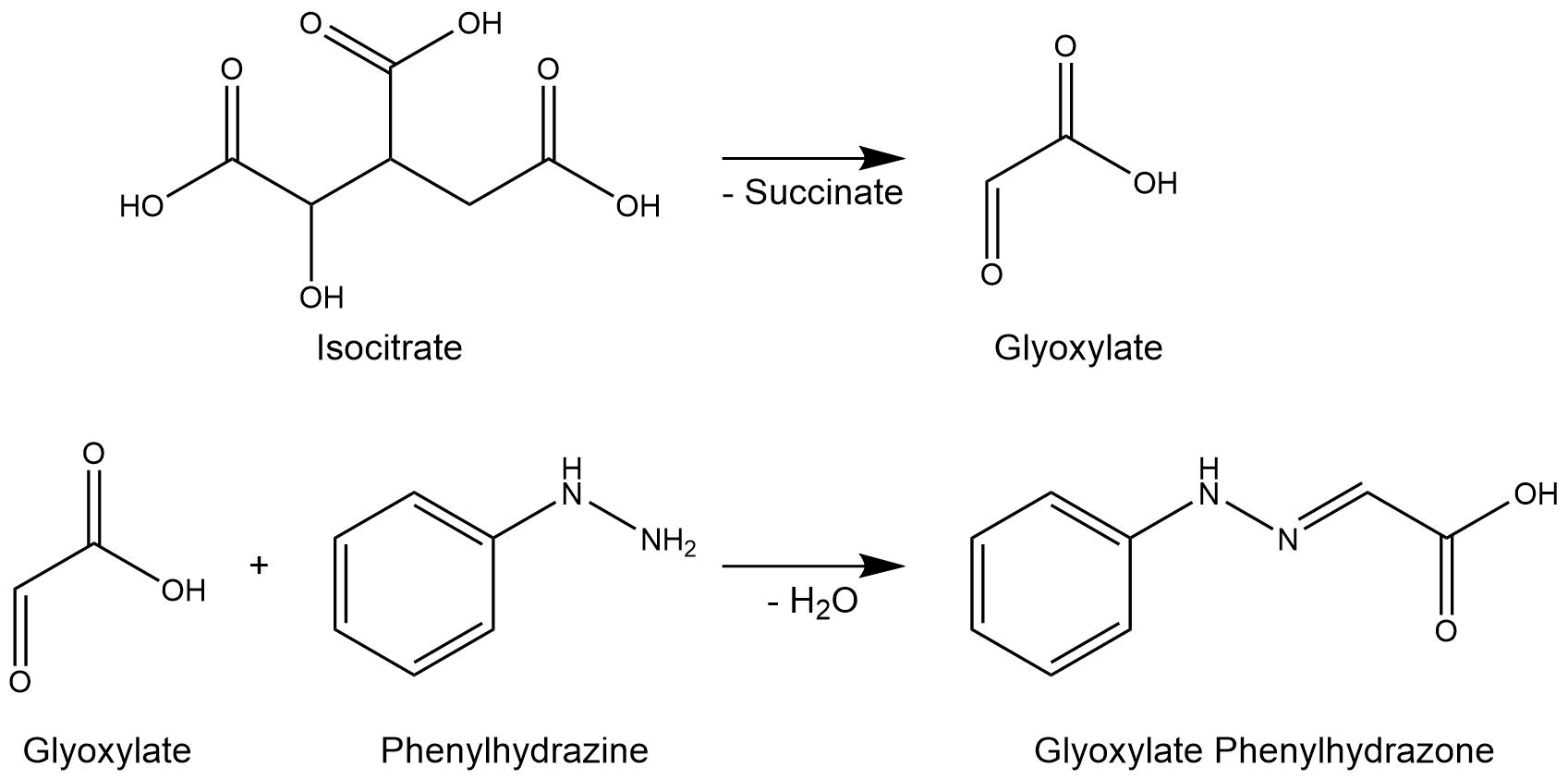
The isocitrate lyase catalyzes the conversion of isocitrate into glyoxylate and succinate. [1]. AceA has a tetrameric structure with a molecular mass of 47.522 kDa. (paper aceA) Bild Enzym aceA
Subsequently, the glyoxylate produced by the isocitrate lyase AceA is further converted into glycolic acid by the NADPH-dependent glyoxylate reductase YcdW. It has molecular weight of 35.343 kDa. Bild Enzym ycdW
Through overexpression of both key enzymes, we achieved the heterologous production of glycolic acid in E. coli. We assess our accomplishment as a crucial step towards a more environment-friendly production of biodegradable polymers. Reaktionsmechanismus
Mechanism
Methods
Cloning
The sequences for aceA and ycdW were modified with a His-tag, ordered from Integrated DNA Technologies (IDT) and inserted into the pSB1C3 plasmid. For this purpose, the BioBrick assembly (BBa) was used. A T7 lac promotor (BBa_K921001) and a B0034-based ribosomal binding site (BBa_K2380024) were inserted upstream of the coding sequence via the BBa as well. E. coli TOP10 were transformed with generated plasmids and positive colonies were identified via colony PCR and DNA sequencing.
 [[Plasmidkarten -> 2 pro Teamseite]]
[[Plasmidkarten -> 2 pro Teamseite]]
SDS-PAGE and Western Blot
To verify that aceA and ycdW were expressed and the corresponding proteins were translated, a SDS-PAGE and Western Blot were performed. The resulting bands were compared to the expected protein sizes of AceA and YcdW.
Purification
After expression of AceA and YcdW in E. coli BL21 by induction of the T7 lac promotor with IPTG, an ÄKTA chromatography system (GE Healthcare, Illinois, USA) was used to purify the desired His-tagged enzymes.
Enzyme assays
The purified enzymes were spectrophotometrically assayed for their activity using a plate reader. For the assay of the isocitrat lyase (AceA), the enzyme activity was tested via a phenylhydrazin-dependent reaction. As soon as AceA converts isocitrate into glyoxylate, the product reacts with phenylhydrazin to glyoxylate-phenylhydrazon, which possesses an absorption maximum at 324 nm. The change in absorption at 324 nm was measured over time.
The assay for the glyoxylate reductase YcdW is based on the different absorption maxima of NADPH (absorption maximum at 340 nm) and NADP (maximum at 260 nm). During the reaction the enzyme uses NADPH as a cofactor. NADPH is converted into NADP which leads to a decrease of absorption at 340 nm. By measuring the absorption level over time, it is possible to quantify the enzyme activity and glycolic acid production.
HPLC analysis
To detect the precursors, as well as the produced monomer glycolic acid, high-performance liquid chromaography (HPLC) was employed. An organic acid separation column as stationary phase and sulfuric acid as mobile phase allowed the separation of isocitrate, glyoxylate and glycolic acid. Signals were recorded by a refractive index detector.
M9 assay
The growth of E. coli BL21 transformed with an YcdW-generating plasmid was studied in M9 minimal media with and without a supplemented carbon source. M9 minimal medium consists of numerous salts to achieve an isotonic environment, but no carbon source. Hence, bacterial growth in M9 minimal media is inhibited. However, by supplementing either glucose, glyoxylate or glycolic acid in M9 media, the potential metabolization of these compounds of interest as a carbon source can be investigated. Therefore, the OD600 of E. coli cultures with the different carbon sources of interest were measured over a period of four days and compared to a carbon source-deficient culture.
Results
Discussion and Outlook
- ↑ CITE ME paper aceA/uniprot)