Background
KcsA
KcsA is a prokaryotic potassium channel from the soil bacteria Streptomyces lividans that has been studied extensively in ion channel research. The pH activated protein possesses two transmembrane segments and a highly selective pore region, responsible for the gating and shuttling of K+ ions from the extracellular environment into the cell. The amino acid sequence (TVGYG) found in the selectivity filter of KcsA is highly conserved among both prokaryotic and eukaryotic K+ voltage channels; as a result, research on KcsA has provided important structural and mechanistic insight on the molecular basis for K+ ion selection and conduction. As one of the most studied ion channels to this day, KcsA is a template for research on K+ channel function and its elucidated structure underlies computational modeling of channel dynamics for both prokaryotic and eukaryotic species[1].
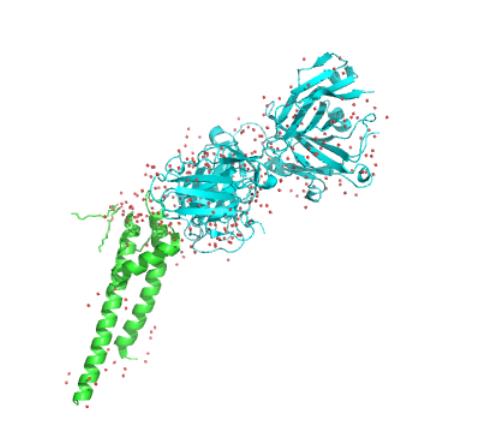
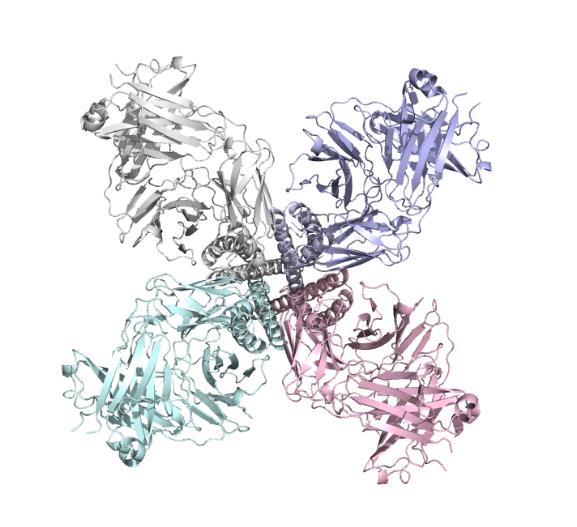

The selective filter is the central structure of the ion channel. Its amino acid sequence is TVGYG from77-79[2], which is the same in all potassium channels. The carbonyl oxygen atoms on the amino acid sequence point inward toward the ion channel and form four ion-binding sites on the potential energy.

[1] Wikipedia
[2] Benoît Roux Ion channels and ion selectivity Essays in Biochemistry Essays Biochem. 2017 May 09; 61(2): 201–209
tRNA
Transfer ribonucleic acid: TRNA refers to a small class of RNA carrying and transporting amino acids. Most tRNAs consist of short clover-shaped strands of more than 70 to 90 nucleotides folded into ribosomes. The main function of tRNA is to carry amino acids into ribosomes and synthesize proteins under the guidance of mRNA. In other words, they translate the sequence of coding nucleotides into the sequence of amino acids in proteins using mRNA as a template. TRNA and mRNA are interacted by anticodon and codons. In the process of peptide chain formation, the first tRNA that enters the ribosome to bind to the mRNA initiation codon is called the initiation tRNA and the remaining tRNA that is involved in the peptide chain extension is called the extension tRNA. According to the sequence of codons on the mRNA, the tRNA carrying specific amino acids enters the ribosome in turn. After forming the peptide chain, tRNA is released from ribosomes. The whole process is called the tRNA cycle. A tRNA can carry one amino acid, such as alanine tRNA, but an amino acid can be carried by more than one tRNA.

aaRS
Aminoacyl-tRNA synthetases: Amino acid activator is an enzyme for the synthesis of aminoacyl tRNA. 20 natural amino acids have their specific specific aminoacyl tRNA synthetase. It is generally believed to participate in the two-step reaction shown below, activating amino acids in the presence of ATP and binding to the - CCA - OH terminal of tRNA. Then, with the help of various translation factors and combinations, it enters the active sites between ribosome size subunits, forms peptide bonds with the last amino acid, and encodes them into proteins. This enzyme can identify the side chains and tRNA of amino acids. Because enzymes have such a highly specific substrate, the genetic information of the mRNA can be accurately reflected in the amino acid sequence of the protein.

Alr
Alanine racemase: Racemase is a kind of isomerase. It can catalyze the transformation between optical isomers containing an asymmetric carbon atom compound. Because the amino acid is alanine, it is called alanine racemase. In our project, it was used to introduce D-alanine into bacteria. Bacteria can't form D-alanine in the body in normal circumstances, but it can convert L-alanine to D-alanine for the purpose.
[1] baidubaike
[2] Nediljko Budisa Prolegomena to Future Experimental Efforts on Genetic Code Engineering by Expanding Its Amino Acid Repertoire Angew. Chem. Int. Ed. 2004, 43, 6426 – 6463

